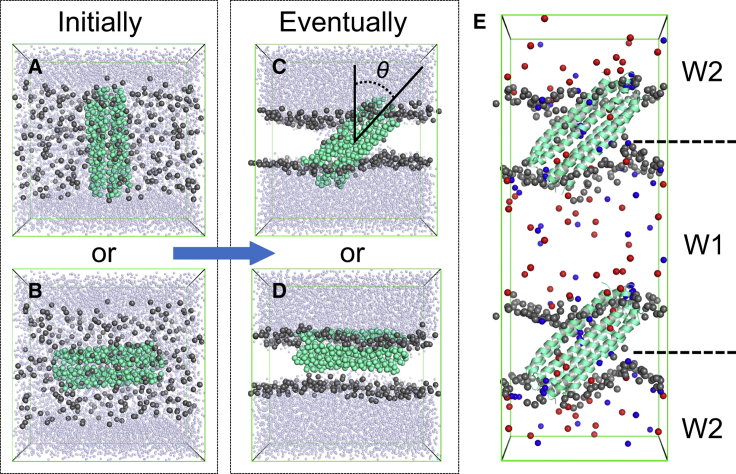
Figure 1.
Coarse-grained (CG) self-assembly simulations of DCD/lipid bilayer systems and the atomistic computational electrophysiology (CE) simulation system. (A) and (B) show two initial systems for the CG simulations, with the helix bundle (green-cyan) perpendicular or parallel, respectively, to the plane of the initial slab containing lipid molecules (dark gray). (C) and (D) show two possible resultant orientations of the DCD oligomer relative to the self-assembled bilayer (lipid phosphate particles only are shown in dark gray) at the end of the 0.5-μs CG simulations. The tilt angle is indicated by θ in (C). The small transparent light-blue spheres are water particles. (E) The atomistic CE simulation system is shown in which the green-cyan cartoons represent the DCD oligomer, the gray spheres are the phosphorus atoms of the lipid (DMPC) headgroups, and the red and blue spheres are Cl− and Na+ ions, respectively. W1 and W2 represent the two aqueous compartments between which a voltage difference of ∼250–450 mV was maintained in the CE simulations. To see this figure in color, go online.