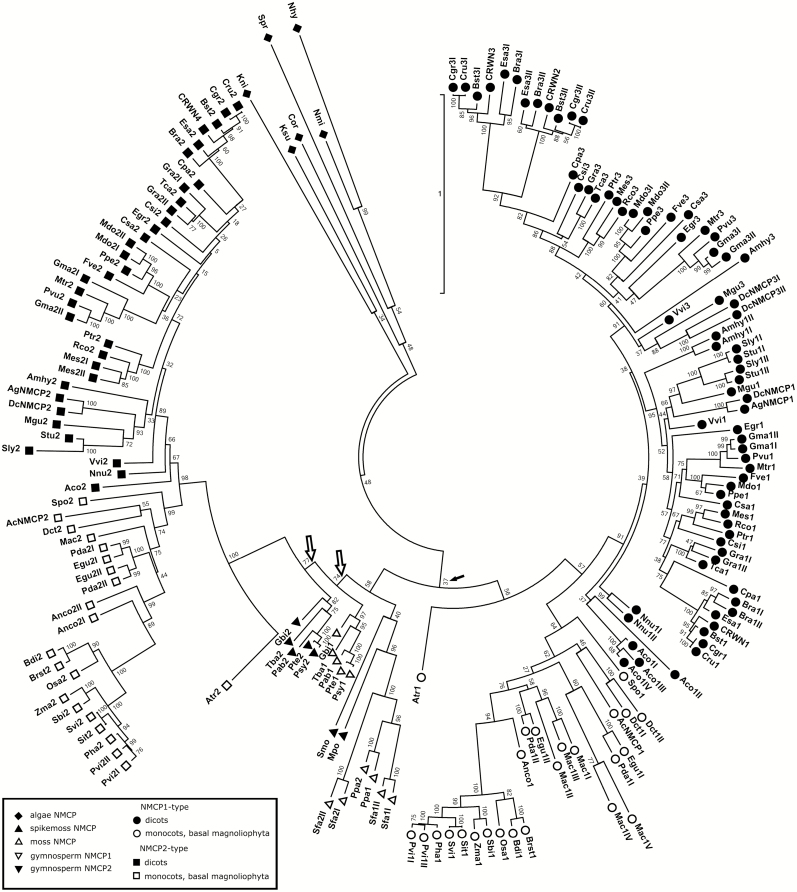
Fig. 2.
Phylogenetic tree of NMCPs. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The tree with the highest log likelihood is shown. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 176 amino acid sequences. There were a total of 2761 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Reliability of the branches was inferred from bootstrap analyses of 1000 replicates. All positions with less than 95% site coverage were eliminated (i.e. less than 5% alignment gaps, missing data or ambiguous bases were allowed at any position). Filled arrows indicate low bootstrap values for the node including NMCP2 and basal NMCPs. Empty arrows indicate the two nodes of gymnosperm NMCP1 and NMCP2.