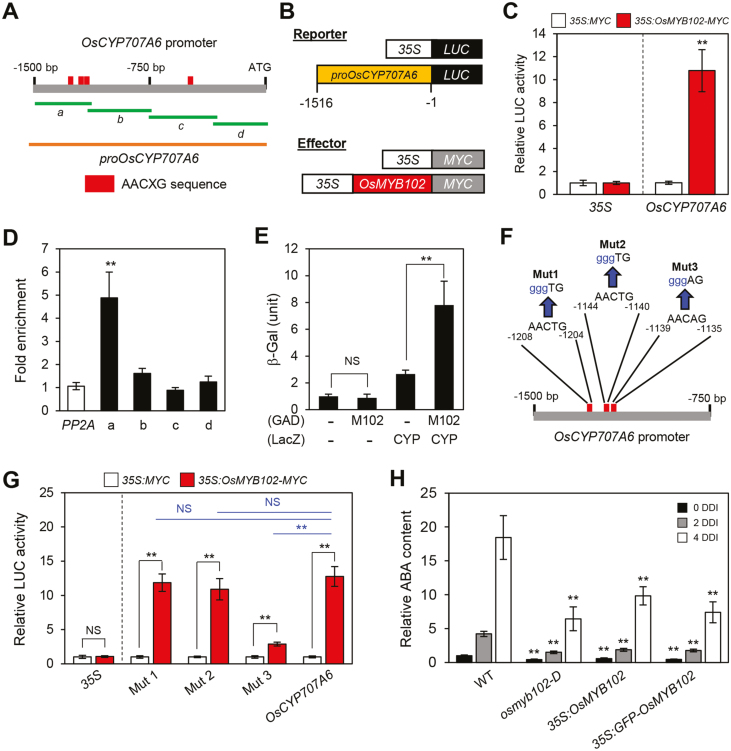
Fig. 6.
OsMYB102 directly activates the transcription of OsCYP707A6. (A) The positions of the AACXG binding motifs in the promoter of OsCYP707A6 and promoter fragments used for the ChIP assay (green horizontal lines), the transactivation assay, and Y1H assay (orange horizontal line). (B) Reporter and effector constructs used in the transactivation assay. Each construct also contains the NOS terminator (not shown). (C) The activation of the OsCYP707A6 promoter (–1516 to –1) by OsMYB102-MYC in the protoplast transient assay. The 35S promoter was used as a negative control. (D) OsMYB102 binding affinity to the promoter region of OsCYP707A6 in planta examined by ChIP assays. OsMYB102-Myc was transiently expressed in protoplasts isolated from 10-day-old WT seedlings. Fold-enrichment of the promoter fragments was measured by immunoprecipitation with an anti-Myc antibody (see Methods). SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PP2A) was used as a negative control. (E) The binding activity of OsMYB102 to the promoter regions of OsCYP707A6 was examined by Y1H assays. Empty bait and prey plasmids (–) were used for the negative controls. The relative β-galactosidase activity was obtained by normalizing to the level of each negative control. The mean and SD values were obtained from more than five independent colonies. (F, G) The effects of disruption of the putative OsMYB102 binding sites in the OsCYP707A6 promoter in rice protoplasts. LUC was fused to the wild-type or mutated OsCYP707A6 promoter fragments, and the OsMYB102 overexpression vector or empty vector were co-transfected in the protoplasts. The 35S promoter was used as a negative control. The mean and SD values were obtained from more than four biological samples. (H) Relative ABA contents in WT, osmyb102-D, 35S:OsMYB102, and 35S:GFP-OsMYB102 plants at 0 (black bars), 2 (gray bars), and 4 d (white bars) after dark incubation. The mean and SD values were obtained from more than five biological samples. (C, D, E, G, H) Asterisks indicate a significant difference compared with the WT or negative control (Student’s t-test, *P<0.05, **P<0.01).