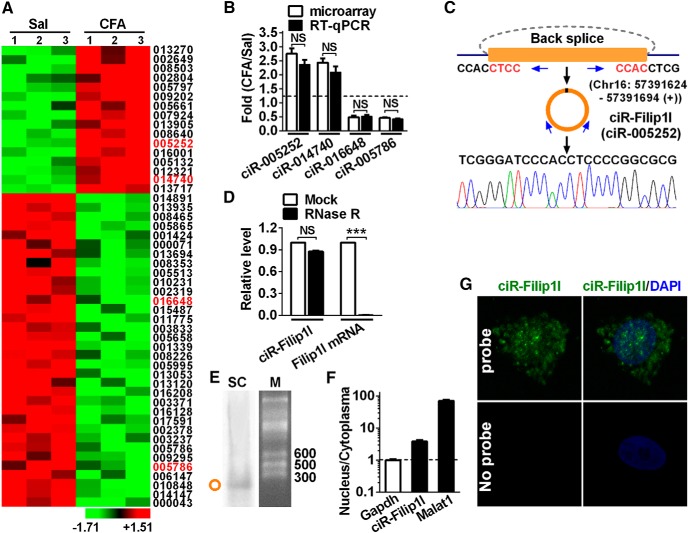
Figure 1.
Profiling of circRNAs and circRNA-Filip1l expression in mouse spinal cord. A, The expression profiling of differential circRNAs with twofold or more was generated from circRNA microarray. Spinal cord was collected 3 d after CFA or saline (Sal) injection. n = 3 per group. The number indicates the identification of circRNA in Arraystar Mouse circRNA's Microarray. B, Four differential circRNAs were subjected to qRT-PCR verification. n = 4 per group. There was no significance (NS) versus the corresponding microarray groups by two-tailed paired Student's t test. C, The genomic loci of circRNA-Filip1l (ciR-Filip1l) is shown. The junction of circRNA-Filip1l was amplified by the use of back-to-back primers and then sequenced by Sanger sequencing. Arrows indicate divergent primers binding to the genome region of circRNA-Filip1l. D, Total RNAs were digested with RNase R followed by qRT-PCR detection of circRNA-Filip1l expression. Filip1l mRNA was detected as the RNase R-sensitive control. n = 4 per group. ***p < 0.001 versus the corresponding mock groups (two-tailed paired Student's t test). E, Northern blot for circRNA-Filip1l in spinal cord of mice. SC, Spinal cord; M, RNA marker. F, Distribution of circRNA-Filip1l in the nucleus and cytoplasma of spinal neuron cultured in vitro. Gapdh, Coding RNA control; Malat1, noncoding RNA control. Their levels were normalized to Gapdh. Spinal nucleus and cytoplasma RNA were collected, respectively, from 48 h cultured mouse spinal neurons in vitro. n = 4 per group. G, circRNA-Filip1l fluorescence in situ hybridization (FISH) in the spinal neuron cultured in vitro. DAPI, Nucleus staining dyes.