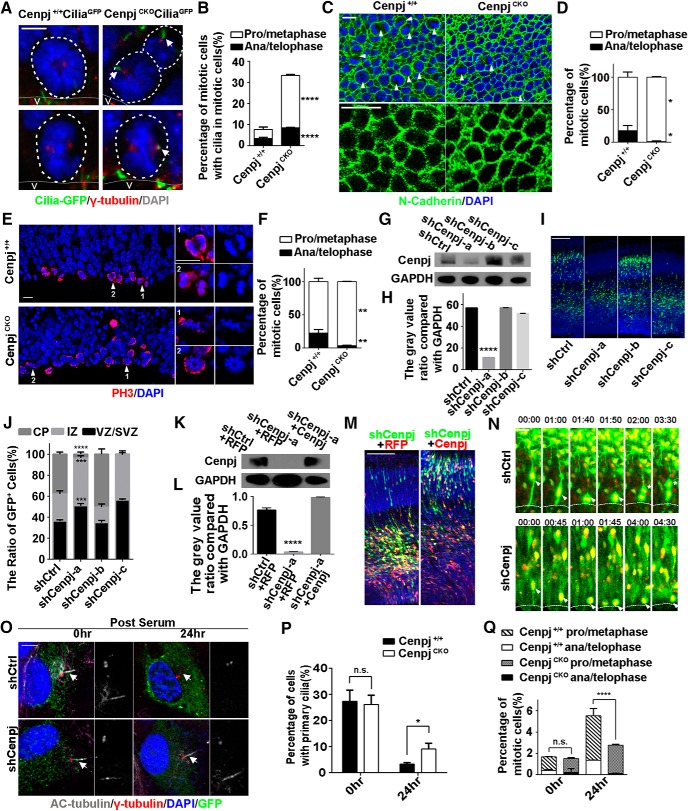
Figure 3.
Cenpj regulates cilia disassembly and RG division. A, Representative images of the mitotic cells on the lateral ventricle surface of Cenpj+/+CiliaGFP and CenpjCKO CiliaGFP embryo brains. Cilia are shown in green and γ-tubulin in red. Scale bar, 10 μm. B, Quantification of the mitotic cells with cilia in prophase/metaphase versus in anaphase/telophase on the Cenpj+/+ and CenpjCKO mouse lateral ventricle surface. Histogram showing the mean ± SD; ****pprophase/metaphase < 0.0001, ****panaphase/telophase < 0.0001 as determined by t test; n = 3; >200 cells per experiment. C, En face views of the embryo ventricular zone in Cenpj+/+ and CenpjCKO mice stained for N-cadherin (green) and DAPI (blue). High-magnification images indicate cell adhesion. Arrowheads indicated the mitotic cells. Scale bar, 10 μm. D, Quantification of the mitotic cells in pro/metaphase versus those in anaphase/telophase on the Cenpj+/+ and CenpjCKO mouse lateral ventricle surface. Histogram showing the mean ± SD; *pprophase/metaphase = 0.0230, *panaphase/telophase = 0.0230 as determined by t test; n = 3; 3 brain slices per experiment. E, Representative images of mitotic cells at the VZ surface in E13.5 Cenpj+/+ or CenpjCKO cortices stained for phosphorylated histone 3 (H3, red) and DAPI (blue). High-magnification images indicate mitotic cells. Scale bar, 10 μm. F, Quantification of the fraction of mitotic cells in prophase/metaphase (white) versus those in anaphase/telophase (black), which reflects mitotic progression. Histogram showing the means ± SD; **pprophase/metaphase = 0.0037, **panaphase/telophase = 0.0037 as determined by t test; n = 3; >200 cells per experiment. G, Cenpj was efficiently knocked down by Cenpj shRNAs. GAPDH serving as a loading control. H, Quantification of Cenpj knockdown efficiency by shRNAs. Histograms show the mean ± SD; ****pshCtrl vs shCenpj-a < 0.0001, as determined by t test; n = 3. I, Analysis of the radial migration of cortices 3 d after IUE at E13.5. Scale bar, 30 μm. J, Quantification of the neurogenesis after silencing Cenpj by measuring the percentages of GFP+ cells that have reached different zones of the cortex 3 d after electroporation. Histograms show the mean ± SD; shCtrl vs shCenpj-a (****pCP < 0.0001, ***pIZ = 0.0008, ***pVZ/SVZ = 0.0004 as determined by a t test; n = 3; 9 brain slices per experiment). K, Cenpj was specifically knocked down by shCenpj-a and was rescued by the overexpression of Cenpj-td plasmid. GAPDH serving as a loading control. L, Quantification of Cenpj protein expression index. Histograms show the mean ± SD; ****pshCtrl+RFP vs shCenpj-a+RFP < 0.0001 as determined by a t test; n = 3. M, Analysis of the radial migration of cortices 3 d after IUE at E13.5. Scale bar, 30 μm. N, Representative time-lapse images of the RG dividing process in the sections of the cerebral cortex electroporated with shCtrl and shCenpj in the wild-type mouse. Arrowheads indicate mother RG cells and asterisks indicate two daughter cells. Scale bar, 20 μm. O, Ciliogenesis in ARPE19 cells. Cells were treated, fixed at 0 h or at 24 h after serum restimulation, and immunostained with anti-γ-tubulin (red) and anti-acetylated tubulin (gray) antibodies. Enlarged views show the centrosome (right top) and the primary cilia (right bottom). Arrows indicate the cilia. Scale bar, 10 μm. P, Percentage of ARPE19 cells with primary cilia in GFP-positive cells. Histogram show the means ± SD; p0 h = 0.7091, *p24 h = 0.0109 as determined by a t test; n > 200 cells. Q, Quantification of the fraction of mitotic cells in prophase/metaphase versus those in anaphase/telophase. Histogram showing the means ± SD; t test; p0 h,prophase/metaphase = 0.9106, p0 h,anaphase/telophase = 0.4686, ****p24 h,prophase/metaphase < 0.0001, ****panaphase/telophase < 0.0001 as determined by a t test; n > 200 cells.