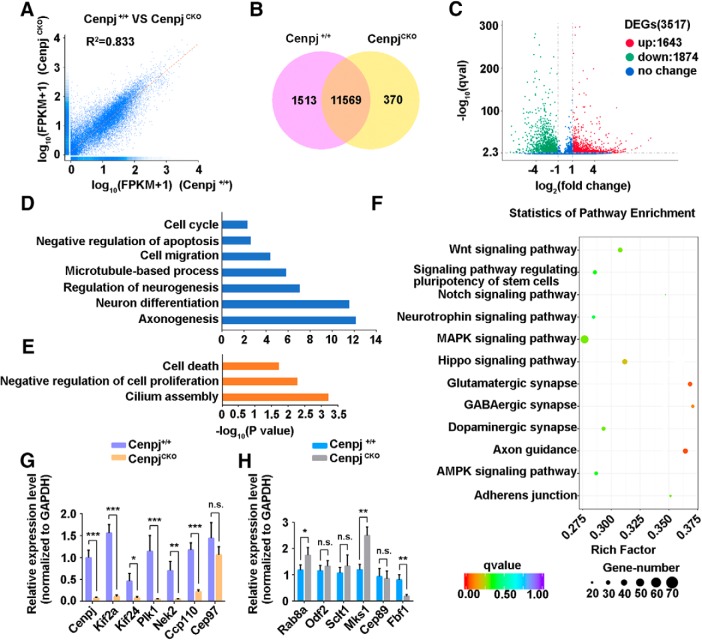
Figure 6.
Transcriptome changes in the developing mouse cortex with Cenpj depletion. A, RNA-Seq correlation of the gene expression level. B, Venn diagrams of the coexpression genes between CenpjCKO samples and Cenpj+/+ samples. C, Scatter plot analysis of transcriptome expression profiles of CenpjCKO samples versus Cenpj+/+ samples at E15.5. Red dots and green dots highlight the significantly upregulated or downregulated expressed genes after Cenpj depletion, respectively. Blue dots show no significantly changed genes. D, Histogram of the enriched gene ontology (GO) terms of downregulated genes from the pairwise comparison of CenpjCKO versus Cenpj+/+. The significance of each GO term was estimated based on corrected p-values (corrected p < 0.05). E, Histogram of the enriched GO terms of upregulated genes from the pairwise comparison of CenpjCKO versus Cenpj+/+. The significance of each GO term was estimated based on corrected p-values (corrected p < 0.05). F, Statistics of enriched KEGG pathway display the varying genes that are involved in many pathways. The size of the point indicates the number of differentially expressed genes in this pathway and the color of the point corresponds to a different q-value range. G, qRT-PCR-analyzed transcription of genes involved in cilium disassembly. All measured values were normalized to GAPDH gene expression. Histogram shows the mean ± SD; ***pCenpj = 0.0008, ***pKif2a = 0.0002, *pKif24 = 0.0220, ***pPlk1 = 0.0002, **pNek2 = 0.0056, ***pCcp110 = 0.0005, pCep97 = 0.1788 as determined by t test; n = 3; 3 repetitions per experiment. H, qRT-PCR-analyzed transcription of genes involved in cilium assembly. All measured values were normalized to GAPDH gene expression. Histogram shows the mean ± SD; *pRab8a = 0.0484, pOdf2 = 0.3714, pSclt1 = 0.3873, **pMks1 = 0.0038, pCep89 = 0.7596, **pFbf1 = 0.0060 as determined by t test; n = 3; 3 repetitions per experiment.