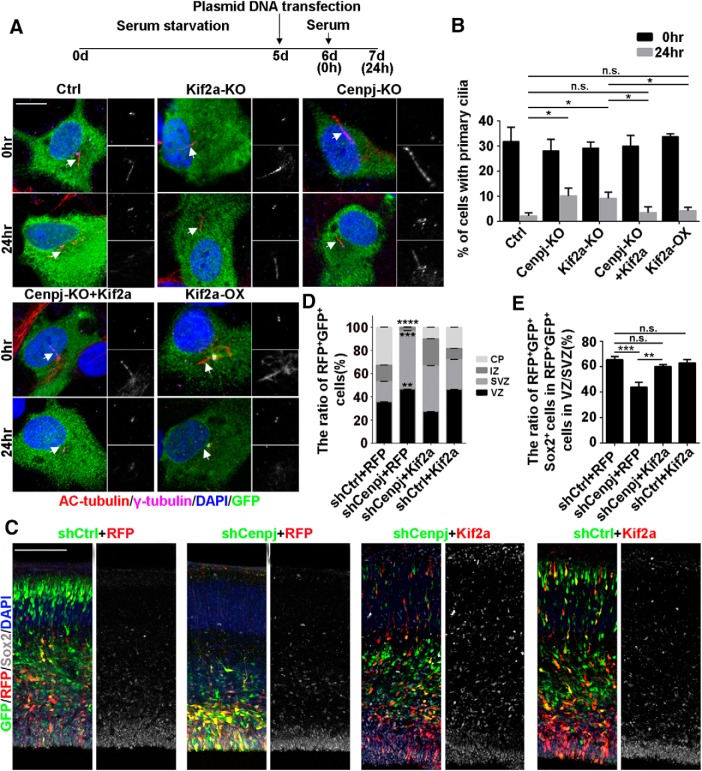
Figure 8.
Kif2a rescues the phenotype caused by Cenpj deletion. A, Ciliogenesis in ARPE19 cells. Cells were treated, fixed at 0 h or at 24 h after serum restimulation, and immunostained with anti-γ-tubulin (purple) and anti-acetylated tubulin (red) antibodies. Enlarged views show the centrosome (right top) and primary cilia (right bottom). Arrows indicate the primary cilia. Scale bar, 10 μm. B, Percentage of ARPE19 cells with primary cilia in GFP-positive cells. Histogram shows the means ± SD; *p24 h, Ctrl vs Cenpj-KO = 0.0151, *p24 h, Ctrl vs Kif2a-KO = 0.0129, p24 h, Ctrl vs Cenpj-KO+Kif2a = 0.4584, p24 h, Ctrl vs Kif2a-OX = 0.1250, *p24 h, Kif2a-KO vs Cenpj-KO+Kif2a = 0.0475, *p24 h, Kif2a-KO vs Kif2a-OX = 0.0424 as determined by a t test; n = 3; >200 cells per experiment. C, Neurogenesis defects caused by Cenpj depletion were rescued by Kif2a expression. Confocal images of cortices 3 d after IUE at E13.5. shRNA-GFP (green), Kif2a-RFP (red), Sox2 (gray). Scale bar, 30 μm. D, Quantification of the migration of neurons by measuring the percentages of GFP+ and RFP+ cells that reached the different zones of the cortex 3 d after electroporation. Histogram shows the means ± SD; shCtrl+RFP vs shCenpj+RFP (****pCP < 0.0001, ****pIZ < 0.0001, ****pSVZ < 0.0001, **pVZ = 0.0039 as determined by a t test; n = 3; 9 brain slices per experiment). E, Quantification of the neural stem cell number was measured by counting the percentage of GFP+RFP+Sox2+ cells in the VZ/SVZ. Histogram shows the means ± SD; ***pshCtrl+RFP vs shCenpj+RFP = 0.0008, pshCtrl+RFP vs shCenpj+Kif2a = 0.5416, pshCtrl+RFP vs shCtrl+Kif2a = 0.8628, **pshCenpj+RFP vs shCenpj+Kif2a = 0.0011, t test; n = 3; 9 brain slices per experiment.