Abstract
The talin proteins are a key component of the extracellular matrix-integrin-cytoskeleton system, and our previous study suggested that talin2 contributes to the tumor invasion and metastasis processes regulated by the tumor microenvironment. In the present study, the specific effects of talin2 on the invasive ability of breast cancer cells, as well as the underlying mechanism, were investigated by creating two MDA-MB-231 cell lines with stable talin2 knockdown by specific RNA interference. Initially, it was confirmed that the expression levels of talin2 in human breast cancer tissues were upregulated compared with in normal adjacent tissues. Subsequently, invasion and wound healing assays revealed that depletion of talin2 in MDA-MB-231 cells decreased their migratory and invasive abilities. Western blot analysis demonstrated that knockdown of talin2 in MDA-MB-231 cells caused marked downregulation of the tumor microenvironment markers hypoxia-inducible factor 1α, phosphorylated ribosomal protein S6 kinase, phosphorylated protein kinase B and phosphorylated mechanistic target of rapamycin. Furthermore, knockdown of talin2 decreased the basal contents of glucose and lactic acid in the breast cancer cell line. In conclusion, the findings of the present study demonstrated that talin2 knockdown may inhibit the invasive ability of human breast cancer MDA-MB-23l cells via alterations in the tumor microenvironment.
Keywords: talin2, tumor microenvironment, breast cancer, invasion
Introduction
Breast cancer is the most common cancer in women worldwide with more than 2 million new cases expected to be diagnosed in 2018, accounting for 25% of all new cancer cases in women. Breast cancer also is the leading cause of cancer death among women worldwide with an estimated 626,700 breast cancer deaths in women in 2018 (1). Breast cancer can be divided into four subtypes that have distinct prognostic and therapeutic implications. Most breast cancers are luminal A (HR+/HER2-), being slow-growing and less aggressive than other subtypes, and with the most favorable prognosis in part due to increased responsiveness to anti-hormone therapy. Triple negative (HR-/HER2-) subtype (TNBC) represents approximately 12% of all breast cancers, with a poorer short-term prognosis than other subtypes. Therefore, there is an urgent need for a greater understanding of TNBC at all levels and for the development of better therapies (2). As a TNBC cell line, MDA-MB-231 cell, distinguished by its invasive phenotype, was always used in the identification of genes and pathways that are potential mediators of metastasis (3).
The tumor microenvironment can markedly affect the invasion and metastasis of cancer cells (4). The main component of the tumor microenvironment is the extracellular matrix, a network of proteins that interact with cell surface receptors to mediate the actin cytoskeleton, and that also contains other signaling molecules, including growth factors and cytokines (5). Talins are large adaptor proteins responsible for the linkage between the integrin family of adhesion molecules and F-actin within cells (6). In vertebrates, two talin genes, TLN1 and TLN2, that encode very similar proteins, talin1 and talin2, have been identified (7,8). Previous studies have established that talin1 can regulate focal adhesion dynamics, cell migration and cell invasion (9–11), and recent studies involving talin2 knockdown have indicated that talin2 serves a role in the migration of hepatocellular carcinoma cells and breast cancer cells (12,13). However, the role of talin2 in cell invasion and metastasis remains unclear. In the present study, talin2 expression was downregulated in MDA-MB-231 human breast cancer cells by short hairpin RNA (shRNA)-mediated interference. Using the established talin2 knockdown cell lines, the effects of loss of talin2 on MDA-MB-231 cell invasion were investigated, along with the influence of the tumor microenvironment on the occurrence and development of breast cancer.
Materials and methods
Patients and samples
A total of 32 breast cancer tissue specimens from tumor and adjacent tissues were obtained from previously untreated female patients. The patients were aged 36–73 years, with a mean age of 53 years, and underwent surgical treatment at the Department of General Surgery, The First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between January 2016 and June 2017. All samples were cut into two samples. One tissue sample was fixed with 4% paraformaldehyde for 24 h at 4°C, then embedded in paraffin and processed for routine histopathological examination, while the other tissue sample was frozen immediately in liquid nitrogen and stored at −80°C for further studies. For all patients, the diagnosis of breast cancer was based on clinical and histopathological examination. Tumor stage was defined according to the tumor node metastasis (TNM) system by Union Internationale Contre le Cancer (14). Tumor pathological grade was defined based on the World Health Organization classification system (15). The present study was approved by the Board and Ethical Committee of The First Affiliated Hospital of Wenzhou Medical University. All study participants provided written informed consent in accordance with the Declaration of Helsinki.
Materials
Anti-talin2 antibody (cat. no. MCA4771GA) was purchased from Bio-Rad Laboratories, Inc. (Hercules, CA, USA). Anti-protein kinase B (AKT; cat. no. 2966S), anti-phosphorylated AKT (pAKT; cat. no. 4060S), anti-mechanistic target of rapamycin (mTOR; cat. no. 2983S), anti-phosphorylated mTOR (pmTOR; cat. no. 5536S), anti-hypoxia-inducible factor 1α (HIF-1α; cat. no. 36168S), ribosomal protein S6 kinase (S6K; cat. no. 2708S), anti-phosphorylated ribosomal protein S6 kinase (pS6K; cat. no. 9204S), anti-tubulin (cat. no. 2148S), anti-actin (cat. no. 3700S) and anti-GAPDH antibodies (cat. no. 2188S) were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (cat. no. A0208), the 3,3′-diaminobenzidine color development kit and the bicinchoninic acid (BCA) protein concentration detection kit (cat. no. P0011) were obtained from Beyotime Institute of Biotechnology (Haimen, China). Talin2 shRNA clones TRCN0000122990 (shRNA1#) and TRCN0000122992 (shRNA2#) were purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Lipofectamine® 2000 was purchased from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco; Thermo Fisher Scientific, Inc. West Pico enhanced chemiluminescence (ECL) reagent was purchased from Pierce; Thermo Fisher Scientific, Inc. Penicillin G and streptomycin were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The glucose assay kit (cat. no. F006) and lactate dehydrogenase (LDH) assay kit (cat. no. A020-2) were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Tissue immunohistochemistry (IHC)
Representative tumor and adjacent normal regions were marked on the paraffin-embedded blocks. Paired cancer and normal cores were punched from a donor block to generate tissue microarray (TMA) blocks. IHC was performed as described previously (16). Briefly, 4-µm TMA slides were baked for 1 h at 60°C, deparaffinized, dehydrated and treated with citrate buffer (pH 6.0) at 100°C for 30 min. The TMA slides were then incubated with 3% hydrogen peroxide at room temperature for 1 h, followed by overnight incubation with anti-talin2 antibodies (1:25 dilution) at 4°C. Subsequently, the slides were incubated with a secondary antibody for 30 min at 37°C (goat anti-rabbit immunoglobulin G HRP-conjugated antibody; 1:2,000 dilution). Subsequently, the slides were incubated with peroxidase-conjugated streptavidin complex reagent and developed with 3,3-diaminobenzidine solution until the desired staining was achieved (4–5 min). Slides were counterstained with hematoxylin, dehydrated, cleared, mounted and examined under a Nikon 80i microscope. The distribution area of talin2 expression in different tissues was analyzed using ImagePro Plus software 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Cell culture and talin2 knockdown with shRNA
MDA-MB-231 human breast cancer cells and 293T cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in DMEM containing 10% FBS, penicillin (100 U/ml) and streptomycin (100 mg/ml) at 37°C in a humidified incubator with 5% CO2 atmosphere. To construct the recombinant lentivirus, VSV-G-pseudotyped lentiviral vectors were produced by co-transfecting 6×106 293T cells with 2 pmol control shRNA (Sigma-Aldrich; Merck KGaA), talin2 shRNA1# or shRNA2#, along with 1 pmol pMDLg.RRE, 0.5 pmol pMD2.G and 0.5 pmol pRSV.REV (Addgene Inc., Cambridge, MA, USA) using Lipofectamine® 2000, according to the manufacturer's protocol. After 8 h, the original medium was replaced with fresh medium, and the lentiviral supernatant was collected at 48 and 72 h post-transfection. The collected supernatant was filtered through a cellulose acetate membrane (0.45-µm pore size).
A total of 1×105 MDA-MB-231 cells were plated into each well of a 6-well plate and infected with 100 µl lentivirus (MOI, 20) for 24 h. Cells that stably expressed lentiviral shRNAs were obtained by selection with 1 mg/ml puromycin over 3–4 weeks at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from MDA-MB-231 and MDA-MB-231 Talin2 knockdown cells using the PureLink RNA kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized at 42°C for 50 min using the SuperScript First Strand Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) using 0.5–1.0 µg RNA according to the manufacturer's protocol. qPCR reactions were performed using SYBR Green PCR master mix reagents (cat. no. 4309155; Invitrogen; Thermo Fisher Scientific, Inc.) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The relative quantification of gene expression for each sample was analyzed using the 2−ΔΔCq method (17). The following primers were used: Talin2 forward, 5′-GGGGAATGTGTGGGGATTGCATCC-3′ and reverse, 5′GATGAGGCGATGCGGCAGGCA-3′; and 18S rRNA forward, 5′-ACCTGGTTGATCCTGCCAGT-3′ and reverse, 5′-CTGACCGGGTTGGTTTTGAT-3′. The thermocycling conditions were as follows: 20 sec at 95°C; followed by 40 cycles of 5 sec at 95°C and 30 sec at 60°C. Each experiment was repeated three times in duplicate.
Cell invasion assay
A total of 100 µl Matrigel (1:30 dilution in serum-free DMEM) was added to each Transwell polycarbonate filter (diameter, 6 mm; pore size, 8 µm; Costar; Corning, Inc., Corning, NY, USA) and incubated at 37°C for 6 h. MDA-MB-231 cells were trypsinized, washed three times with DMEM containing 1% FBS, and resuspended in DMEM containing 1% FBS at a density of 5×105 cells/ml for seeding into the upper chamber (100 µl/well). Subsequently, 600 µl DMEM containing 10% FBS was added to the lower chambers. As described previously (18), cells were allowed to invade for 36 h in a CO2 incubator at 37°C. The invaded cells were fixed for 15 min with 3.7% formaldehyde and stained using 0.1% crystal violet in 10% ethanol for 30 min at room temperature. The number of invaded cells per field was counted under a light microscope at ×400.
Wound healing assay
Once cells reached 100% confluence in 6-well culture plates, an artificial ‘wound’ was created using a 10-µl pipette tip to scratch the cell monolayer. The wound area was inspected after 36 and 48 h on an inverted phase-contrast microscope with a digital camera at room temperature. The wound healing speed was calculated as the percentage of the initial wound area that remained at different time points until total wound closure.
Western blot analysis
A total of 2×106 cells (MDA-MB-231 or MDA-MB-231 talin2 knockdown cells) were seeded overnight in 6-well plates, prior to being washed with ice-cold PBS and harvested using radioimmunoprecipitation assay buffer (cat. no. P0013C; Beyotime Institute of Biotechnology). The protein concentration was determined using the BCA method, and 20 µg total proteins were separated using 12% SDS-PAGE prior to western blot analysis. The proteins were transferred to a nitrocellulose membrane that was blocked in 5% milk for 1 h at room temperature. The expression levels of AKT, pAKT, mTOR, pmTOR, HIF-1α, total S6K, pS6K, GAPDH and actin were determined by incubating the membrane with the specific antibodies (1:1,000 dilution in 5% milk) overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody (1:1,000 dilution) for 1 h at room temperature prior to being developed with West Pico ECL reagent.
Measurement of glucose and LDH
According to the manufacturer's protocols, 1×107 cells were cultured in 6-well plates for 24 h prior to treatment with 1% Triton X-100 for 30 min. The glucose concentration of the cultured cells was determined using a Glucose Assay Kit. The amount of lactic acid was measured with an Lactate Dehydrogenase Assay Kit. In this assay, LDH catalyzes the synthesis of pyruvic acid from lactic acid and pyruvic acid reacts to form 2,4-dinitrophenyl-hydrazine, which has a visible brownish red color. Following the reaction, the absorbance was read at a wavelength of 490 nm on a microplate reader (Varioskan Flash; Thermo Fisher Scientific, Inc.).
Statistical analysis
All data are presented as the means ± standard deviation and analyzed using single-factor analysis of variance. Multiple comparisons among groups were performed using one-way ANOVA followed by the least significant difference post hoc test for comparison between two groups. The software package SPSS version 19.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analyses. The results are representative of three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
Talin2 expression in breast cancer tissue
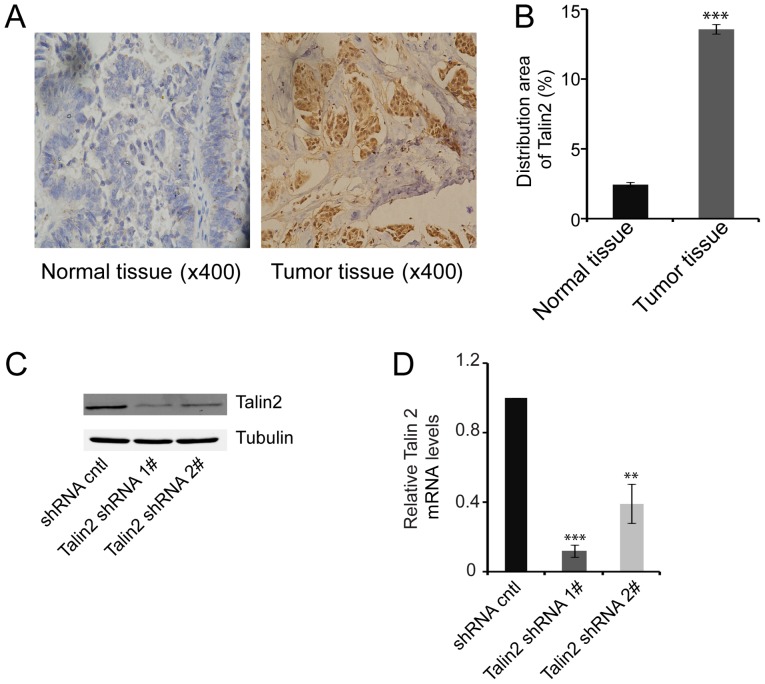
IHC staining for talin2 in breast cancer tissues and adjacent normal tissue samples revealed that talin2 expression was 5.6 times higher in breast cancer tissues compared with in normal tissues (Fig. 1A and B), indicating that talin2 expression may promote the development of breast cancer.
Figure 1.
Expression of talin2 in MDA-MB-231 cells and breast cancer tissues. (A) Distribution of talin2 expression in tumor tissues and normal tissues based on IHC (magnification, ×400). (B) Semi-quantification of IHC results. ***P<0.001 compared with normal tissues. (C) Endogenous talin2 protein levels in talin2-depleted and control MDA-MB-231 cells, as measured by western blotting. (D) Relative mRNA expression levels of talin2 in talin2-depleted and control MDA-MB-231 cells, as measured by quantitative polymerase chain reaction. **P<0.01, ***P<0.001 compared with shRNA cntl. cntl, control; IHC, immunohistochemistry; shRNA, short hairpin RNA.
Confirmation of talin2 knockdown in cancer cells
Following infection of MDA-MB-231 cells with one of the two recombinant talin2 shRNA lentiviruses, the expression levels of talin2 in the two knockdown cell lines were significantly lower compared with in the control group, as detected by western blotting and quantitative polymerase chain reaction (Fig. 1C and D). The talin2 mRNA expression levels in control cells were ~5 or 2 times greater than those in the talin2 shRNA1 and shRNA2 knockdown cell lines, respectively (Fig. 1D). These results indicated that two stable talin2 knockdown cell lines were established, and these lines were used in subsequent experiments.
MDA-MB-231 cell invasion and migration following talin2 knockdown
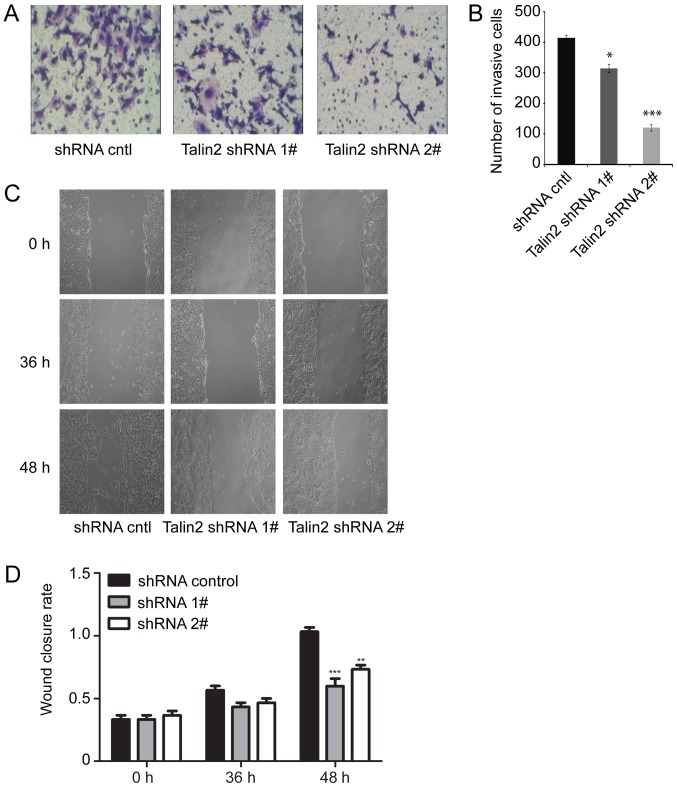
To examine the role of talin2 in breast cancer cell invasion, the invasion of MDA-MB-231 cells infected with talin2 shRNAs or a control shRNA was compared by Transwell assay. The results demonstrated that invasion of MDA-MB-231 cells was significantly reduced by talin2 knockdown compared with in the control group. Specifically, the numbers of invading talin2 shRNA1 and shRNA2 knockdown cells were only 72 and 25%, respectively, of the number for control cells (Fig. 2A and B). Therefore, depletion of endogenous talin2 inhibited the invasive ability of MDA-MB-231 cells.
Figure 2.
Talin2 knockdown inhibits MDA-MB-231 cell invasion and migration. (A) Representative images of invading MDA-MB-231 cells in Transwell assay (magnification ×400). (B) Quantification of invasion assay data. (C) Representative images from wound healing assay (magnification ×200). (D) Quantification of wound healing assay data. *P<0.05, **P<0.01, ***P<0.001 compared with shRNA cntl. cntl, control; shRNA, short hairpin RNA.
To assess whether talin2 regulated the migration of breast cancer cells, an artificial ‘wound’ was created in monolayers of cultured cells using a 10-µl pipette tip, and the rate at which cells grew into the wound area was examined after 36 and 48 h (Fig. 2C and D). After 36 h, the wound healing rate of the talin2 shRNA1 or talin2 shRNA2 knockdown cells was slightly slower compared with in the control group, and after 48 h, the two talin2 knockdown cell lines exhibited slower wound healing (0.67 times and 0.75 times for talin2 shRNA1 and shRNA2 knockdown cells, respectively) than the control cells (P<0.01). These results indicated that talin2 depletion reduced the migratory ability of the breast cancer cells, which was similar to the results of Liang et al (13).
Effect of talin2 knockdown on AKT signaling in the tumor microenvironment
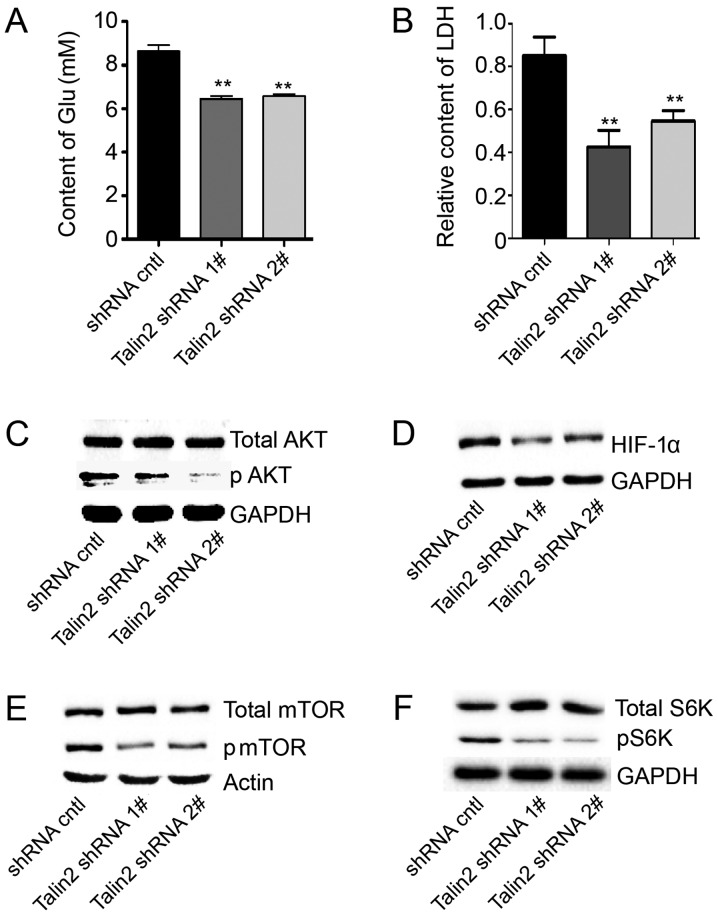
The tumor microenvironment is essential to cancer cell survival and serves an important role in tumor growth and metastasis. Basal glucose and LDH levels are important indicators of the tumor microenvironment. Therefore, basal glucose and LDH contents in MDA-MB-231 cells with or without talin2 knockdown were assessed. The relative glucose levels in talin2 shRNA1 and shRNA2 knockdown cells were 6.912±0.228 and 6.941±0.156 mM, respectively, which was ~20% less compared with control cells (Fig. 3A). The LDH test demonstrated that the lactic acid content in the talin2 shRNA1 and shRNA2 knockdown cells was 0.425±0.090 and 0.546±0.078, respectively, which was 50 and 36% less than that in control cells (Fig. 3B). These results demonstrated that talin2 knockdown may reduce basal glucose and LDH levels, which may stop tumor metastasis by altering the tumor microenvironment.
Figure 3.
Talin2 knockdown affects glucose and LDH levels via the AKT/mTOR signaling pathway. (A) Glucose contents of talin2-depleted and control MDA-MB-231 cells. (B) LDH levels of talin2-depleted and control MDA-MB-231 cells. Expression levels of (C) total AKT and pAKT, (D) HIF-1α, (E) total mTOR and pmTOR, and (F) total S6K and pS6K in talin2-depleted and control MDA-MB-231 cells, as measured by western blotting. **P<0.01, compared with shRNA cntl. AKT, protein kinase B; cntl, control; Glu, glucose; HIF-1α, hypoxia-inducible factor 1α; LDH, lactate dehydrogenase; mTOR, mechanistic target of rapamycin; p, phosphorylated; S6K, ribosomal protein S6 kinase; shRNA, short hairpin RNA.
To further investigate the mechanism by which talin2 knockdown affected the tumor microenvironment, the expression levels of proteins associated with the phosphoinositide 3-kinase (PI3K)/AKT/mTOR signaling pathway, which serves an important role in substance and energy metabolism in the tumor microenvironment, were investigated. Compared with levels in control cells, the expression levels of pAKT and pmTOR in the two talin2 knockdown lines were markedly decreased. In addition, pS6K and HIF-1α expression was clearly reduced (Fig. 3C-F). These results demonstrated that talin2 knockdown may inhibit tumor invasion by altering the tumor environment via its effects on AKT/mTOR signaling.
Discussion
In the present study, which investigated the role of talin2 in breast cancer cells, it was demonstrated that talin2 expression was higher in breast cancer tissues compared with in normal adjacent tissues. Cell invasion and migration assays demonstrated that loss of talin2 in two MDA-MB-231 cell lines, following shRNA-mediated talin2 knockdown, resulted in inhibition of cell invasion and migration. Therefore, the results indicated that talin2 may be overexpressed in breast cancer, and may contribute to cancer cell invasion and metastasis. To investigate the underlying mechanism for this role of talin2 in breast cancer, the effects of talin2 knockdown on the expression of key proteins in the tumor microenvironment were explored.
The tumor microenvironment serves an important role in tumor growth and metastasis (19). This special environment contains tumor cells, endothelial cells, extracellular matrix, immune cells and mechanocytes, under conditions of low pH and low oxygen, with characteristic tumor angiogenesis and a high glucose absorption rate (20). Unlike normal cells, tumor cells primarily obtain energy via anaerobic digestion independent of whether sufficient oxygen is present or not, and this is one of the most fundamental alterations in cell metabolism during malignant transformation. Warburg (21) demonstrated that tumor cells require more glucose than normal cells, and glucose is eventually converted into lactic acid via the glycolysis metabolic pathway, which suggests that cancer is a ‘metabolic disease’. This previous study revealed that tumor cells prefer glycolysis to mitochondrial oxidative phosphorylation for the production of adenosine 5′-triphosphate, even when oxygen levels are sufficient; this phenomenon is known as the Warburg effect. Due to the rapid growth and vigorous metabolism of tumors, lactic acid, a metabolite of glycolysis, accumulates, creating a local acidic environment. Within an acidic microenvironment of pH <7.2, normal cells have difficulty surviving, but tumor cells not only adapt to the acidic environment, but exhibit enhanced ability for invasion and metastasis (22). HIF-1α is the central promoter of the hypoxia-adapted tumor response, and it upregulates pyruvate dehydrogenase kinase-1 and inhibits pyruvate dehydrogenase to inhibit the tricarboxylic acid cycle and the oxidative phosphorylation pathway (23). Additionally, the anaerobic glycolysis pathway is activated. Notably, HIF positively regulates the majority of key enzymes in the glycolysis process (24). Specifically, increased HIF-1α expression under hypoxia activates the 5-terminal enhancer sequence of the glucose transporter type 1 (GLUT-1) gene, promoting large increases in GLUT-1 expression and product synthesis (25,26).
Abnormal PI3K/AKT signaling is an important biological characteristic of tumor cells. As a downstream effector of PI3K, AKT is activated by phosphorylation when it is transported from the cytoplasm to the cell membrane. Blocking the PI3K/AKT signaling pathway can induce a specific antitumor effect (27). mTOR is the most important effector molecule downstream of PI3K/AKT, and its activation activates the entire signaling pathway to control cell growth and proliferation, as well as substance and energy metabolism (28,29). A previous study demonstrated that hypoxia can activate PI3K/AKT, mitogen-activated protein kinase and other signal transduction pathways to stimulate increased expression of HIF-1α and contribute to the pathogenesis of various diseases (30). Phosphorylation of AKT promotes the utilization of glucose by cancer cells by stimulating glycolysis (31).
In the present study, the expression of proteins in the AKT/mTOR signaling pathway was detected following talin2 knockdown in MDA-MB-231 cells to explore the mechanism by which talin2 regulated breast cancer cell invasion. Reduced expression of pS6K and HIF-1α, as well as reduced levels of activated AKT and mTOR, were identified in MDA-MB-231 cells with talin2 knockdown. Additionally, tests for basal glucose and LDH indicated that lactic acid and glucose levels were significantly reduced in breast cancer cells following talin2 knockdown. These data demonstrated that talin2 knockdown may affect the tumor microenvironment via downregulation of AKT/mTOR signaling. Thus, these results may provide a novel target and basis for the diagnosis and treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
This study was supported in part by grants from the Natural Science Foundation of Zhejiang Province (grant no. LY15C070003), the National Natural Science Foundation of China (grant no. 81601522), the Natural Science Foundation of Jiangsu Province (grant no. BK20160348), the Zhejiang University Student Science and Technology Innovation Activity Plan and Xinmiao Talents Program (grant nos. 2017R413056 and 2017R413087), and The Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY conceived and designed the experiments. YL, ZW, SD, LZ and XX performed the experiments. JY, ZW, YL and YW analyzed the data. SD and ZW contributed materials and analysis tools. JY wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Board and Ethical Committee of The First Affiliated Hospital of Wenzhou Medical University. All study participants provided written informed consent in accordance with the Declaration of Helsinki.
Patient consent for publication
All patients provided consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.American Cancer Society, corp-author. Global Cancer Facts & Figures. 4th Edition. American Cancer Society; Atlanta, GA: 2018. [Google Scholar]
- 2.American Cancer Society, corp-author. Breast Cancer Facts & Figures 2017–2018. American Cancer Society; Atlanta, GA: 2017. [Google Scholar]
- 3.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung SY, Hsieh CL, Wu D, Chung LW, Johnstone PA. Tumor microenvironment promotes cancer progression, metastasis and therapeutic resistance. Curr Probl Cancer. 2007;31:36–100. doi: 10.1016/j.currproblcancer.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Liu S, Huang J, Guo W, Chen J, Zhang L, Zhao B, Peng J, Wang A, Wang Y, et al. The ECM-cell interaction of cartilage extracellular matrix on chondrocytes. Biomed Res Int. 2014;2014:648459. doi: 10.1155/2014/648459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debrand E, El Jai Y, Spence L, Bate N, Praekelt U, Pritchard CA, Monkley SJ, Critchley DR. Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J. 2009;276:1610–1628. doi: 10.1111/j.1742-4658.2009.06893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senetar MA, Moncman CL, McCann RO. Talin2 is induced during striated muscle differentiation and is targeted to stable adhesion complexes in mature muscle. Cell Motil Cytoskeleton. 2007;64:157–173. doi: 10.1002/cm.20173. [DOI] [PubMed] [Google Scholar]
- 8.Tsujioka M, Yoshida K, Inouye K. Talin B is required for force transmission in morphogenesis of Dictyostelium. EMBO J. 2004;23:2216–2225. doi: 10.1038/sj.emboj.7600238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat Cell Biol. 2009;11:624–630. doi: 10.1038/ncb1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin JK, Tien PC, Cheng CJ, Song JH, Huang C, Lin SH, Gallick GE. Talin1 phosphorylation activates β1 integrins: A novel mechanism to promote prostate cancer bone metastasis. Oncogene. 2015;34:1811–1821. doi: 10.1038/onc.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang KP, Dai W, Ren YH, Xu YC, Zhang SM, Qian YB. Both Talin-1 and Talin-2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC Cancer. 2016;16:45. doi: 10.1186/s12885-016-2076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, Chen H, Ji L, Du J, Xie X, Li X, Lou Y. Talin2 regulates breast cancer cell migration and invasion by apoptosis. Oncol Lett. 2018;16:285–293. doi: 10.3892/ol.2018.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th edition. John Wiley and Sons; 2009. [Google Scholar]
- 15.World Health Organization, corp-author. Tumours of the Breast and Female Genital Organs. Oxford University Press; 2003. [Google Scholar]
- 16.Liu B, Pang B, Hou X, Fan H, Liang N, Zheng S, Feng B, Liu W, Guo H, Xu S, Pang Q. Expression of high-mobility group AT-hook protein 2 and its prognostic significance in malignant gliomas. Hum Pathol. 2014;45:1752–1758. doi: 10.1016/j.humpath.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Li X, Sunkara M, Spearman H, Morris AJ, Huang C. PIPKIγ regulates focal adhesion dynamics and colon cancer cell invasion. PLoS One. 2011;6:e24775. doi: 10.1371/journal.pone.0024775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spano D, Zollo M. Tumor microenvironment: A main actor in the metastasis process. Clin Exp Metastasis. 2012;29:381–395. doi: 10.1007/s10585-012-9457-5. [DOI] [PubMed] [Google Scholar]
- 20.Swartz MA, Iida N, Roberts EW, Sangaletti S, Wong MH, Yull FE, Coussens LM, DeClerck YA. Tumor microenvironment complexity: Emerging roles in cancer therapy. Cancer Res. 2012;72:2473–2480. doi: 10.1158/0008-5472.CAN-12-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 22.Fais S, Venturi G, Gatenby B. Microenvironmental acidosis in carcinogenesis and metastases: New strategies in prevention and therapy. Cancer Metastasis Rev. 2014;33:1095–1108. doi: 10.1007/s10555-014-9531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upadhyay M, Samal J, Kandpal M, Singh OV, Vivekanandan P. The Warburg effect: Insights from the past decade. Pharmacol Ther. 2013;137:318–330. doi: 10.1016/j.pharmthera.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Nozawa-Suzuki N, Nagasawa H, Ohnishi K, Morishige K. The inhibitory effect of hypoxic cytotoxin on the expansion of cancer stem cells in ovarian cancer. Biochem Biophys Res Commun. 2015;457:706–711. doi: 10.1016/j.bbrc.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 25.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 26.Icard P, Lincet H. A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim Biophys Acta. 2012;1826:423–433. doi: 10.1016/j.bbcan.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Gaikwad SM, Ray P. Non-invasive imaging of PI3K/Akt/mTOR signalling in cancer. Am J Nucl Med Mol Imaging. 2012;2:418–431. [PMC free article] [PubMed] [Google Scholar]
- 28.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Coddou C, Sandoval R, Castro P, Lazcano P, Hevia MJ, Rokic M, Hall B, Terse A, Gonzalez-Billault C, Kulkarni AB, et al. Cyclin-dependent kinase 5 modulates the P2X2a receptor channel gating through phosphorylation of C-terminal threonine 372. Pain. 2017;158:2155–2168. doi: 10.1097/j.pain.0000000000001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hixon ML, Paccagnella L, Millham R, Perez-Olle R, Gualberto A. Development of inhibitors of the IGF-IR/PI3K/Akt/mTOR pathway. Rev Recent Clin Trials. 2010;5:189–208. doi: 10.2174/157488710792007329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.