Abstract
Increasing evidence has revealed that microRNAs (miRNAs) are closely associated with multiple myeloma (MM) pathogenesis and progression. Therefore, an in-depth understanding of the biological functions of miRNAs in MM may be helpful for the identification of promising therapeutic techniques for patients with MM. miRNA-765 (miR-765) has been reported to be dysregulated in many types of human cancer. However, the expression pattern, specific roles and underlying mechanisms of miR-765 in MM remain largely unknown. In the present study, plasma miR-765 significantly increased in patients with MM and cell lines. The downregulation of miR-765 in MM cells attenuated proliferation and promoted apoptosis. Bioinformatics analysis predicted that SRY-Box 6 (SOX6) was a putative target of miR-765. This was experimentally verified using a luciferase reporter assay, reverse transcription-quantitative PCR and western blot analysis. Furthermore, plasma SOX6 was downregulated in patients with MM and the downregulation of SOX6 was inversely correlated with that of miR-765 expression. Furthermore, SOX6 knockdown markedly abrogated the effects of miR-765 underexpression on cell proliferation and apoptosis in MM. The current study demonstrated that miR-765 serves an oncogenic role in MM progression by directly targeting SOX6, suggesting that miR-765 may be a potential therapeutic target for MM prevention and treatment.
Keywords: multiple myeloma, microRNA-765, proliferation, apoptosis, SRY-Box 6
Introduction
Multiple myeloma (MM), the second most common hematologic malignant tumor, is characterized by the clonal proliferation and accumulation of plasma cells (PCs) in bone marrow (1). The morbidity of MM is 0.5–1/100,000 in Asia, whereas the morbidity in Africa and America is 10–12/100,000 (2). Currently, immunomodulatory medicine, proteasome inhibitors and autologous stem cell transplantation are primary therapeutic strategies for patients with MM (3). The techniques used for the diagnosis and therapy of patients with MM have recently progressed; however, the clinical outcomes of patients with MM remain poor with a 5-year survival rate of only 30–40% remaining (4). Further research is therefore required to understand the mechanisms underlying MM pathogenesis and to develop novel treatment options for patients with this disease.
microRNAs (miRNAs) are endogenous, non-coding, short RNA molecules that are ~22 nucleotides in length (5). miRNAs are able to silence gene expression by directly interacting with complementary sites within the 3′-untranslated regions (3′-UTRs) of their target gene to cause mRNA degradation and/or translational suppression (6). Each miRNA may regulate hundreds of different genes. Thus implicating the regulation of a wide range of biological processes, including cell proliferation, cycle, apoptosis, invasion, epithelial-mesenchymal transition, metastasis and drug resistance (7–9). Previous studies have disclosed the importance of miRNAs in MM formation and progression (10–12). A variety of miRNAs have been revealed to be dysregulated in MM (13–15). For example, miR-338-3p (16), miR-324-5p (17) and miR-320c (18) are downregulated in MM; whereas miR-19a (19), miR-32 (20), and miR-210 (21) are upregulated in MM. Dysregulated miRNAs may serve as oncogenes or tumor suppressors, thus contributing to MM malignant progression (22,23). Therefore, further clarification concerning the expression pattern, roles and underlying molecular mechanisms of miRNAs in MM would provide novel clinical intervention tools for patients with this fatal malignancy.
miR-765 is reportedly abnormally expressed and contributes to the tumorigenesis of many different types of human cancer, such as esophageal squamous cell carcinoma (24), osteosarcoma (25) and hepatocellular carcinoma (26). However, the expression pattern, specific roles and underlying mechanism of miR-765 in MM remain largely unknown. The expression of miR-765 in MM cell lines and MM patient plasma was detected in the present study. In addition, the detailed roles and associated mechanisms of miR-765 in MM were examined. Data obtained in the present study may aid the elucidation of the functional roles of miR-765 in MM carcinogenesis and progression.
Materials and methods
Clinical specimens
Bone marrow aspirates were collected from 27 MM patients (15 males, 12 females; age range, 23–57 years) and 11 healthy individuals (7 males, 4 females; age range, 36–52 years) at The First Affiliated Hospital of Nanchang University, Nanchang, P.R. China between June 2015 and August 2017. Patients treated with radiotherapy, chemotherapy, immunomodulatory medicine, proteasome inhibitors and autologous stem cell transplantation were excluded from the current study. Plasma cells were purified from bone marrow aspirates using CD138 MicroBeads (cat. no. 130-051-301; Miltenyi Biotec GmbH) in accordance with manufacturer's protocol. Plasma cells were quickly frozen in liquid nitrogen and then maintained at −80°C. The current study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (Nanchang, China). All participants provided written informed consent prior to enrollment.
Cell lines
Three human MM cell lines (U266, MM1S and RPMI-8226) were purchased from the American Type Culture Collection. RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (all, Gibco; Thermo Fisher Scientific, Inc.) was used to culture all MM cell lines. Cells were maintained at 37°C in a humidified chamber supplied with 5% CO2.
Transfection assay
A miR-765 inhibitor (5′CAUCACCUUCCUUCUCCUCCA3′), a corresponding negative control miRNA inhibitor (NC inhibitor; 5′ACUACUGAGUGACAGUAGA3′), a small interfering RNA (siRNA) targeting SOX6 expression (SOX6 siRNA; 5′GCAGGAAUUUGGACACCUU3′) and a negative control siRNA (NC siRNA; 5′UUCUCCGAACGUGUCACGUTT3′) were ordered from Shanghai GenePharma Co., Ltd. Cells were plated into six-well plates at a density of 5×105 cell per well and cultured overnight at 37°C. Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. Reverse transcription-quantitative (RT-q) PCR and flow cytometric analysis of cell apoptosis were performed 48 h following transfection. A Cell counting kit-8 (CCK-8) assay and western blot analysis were conducted at 24 and 72 h post-transfection, respectively.
RT-q PCR
Total RNA was extracted from tissues or cells using a TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used to determine the concentration of total RNA. To detect miR-765 expression, cDNA was synthesized from total RNA using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The temperature protocol for reverse transcription was as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. qPCR was then performed using a TaqMan MicroRNA PCR kit with an Applied Biosystems 7500 Sequence Detection system (each from Applied Biosystems; Thermo Fisher Scientific, Inc.). The temperature protocol for qPCR were as follows: 50°C for 2 min, 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec; and annealing/extension at 60°C for 60 sec. For the measurement of SOX6 mRNA expression, reverse transcription was performed using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.). The temperature protocol for reverse transcription was as follows: 37°C for 15 min and 85°C for 5 sec. Synthesized cDNA was subsequently used for qPCR using a SYBR Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.). The temperature protocol for qPCR was as follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. U6 small nuclear RNA and GAPDH served as an internal control to normalize the relative expression level of miR-765 and SOX6 mRNA, respectively. Relative gene expression was calculated using the 2−ΔΔCq method (27). The following primers were utilized for qPCR: miR-765 forward, 5′-GUAGCCAAGGAATCCGAAGGA-3′ and reverse, 5′-GCGAGGAAGGAGGAGGAAGGT-3′; U6 forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; SOX6 forward, 5′-CCCGTACAGTTCATTCCGTC-3′ and reverse, 5′-AGCCTTGGGTTAATTTGTGG-3′; GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Cell counting kit-8 (CCK-8) assay
Cellular proliferation was determined using a CCK-8 assay (Dojindo Molecular Technologies, Inc.) as per the manufacturer's protocol. Transfected cells were collected at 24 h post-transfection, suspended in RPMI-1640 medium and inoculated into each well of a 96-well plate at a density of 3×103 cells. Cells were then incubated at 37°C with 5% CO2 at different time points (0, 24, 48 and 72 h). A total of 10 µl CCK-8 solution was added into each well and incubated for 2 h at 37°C. Absorbance at 450 nm was determined using a microplate reader (Bio-Rad Laboratories, Inc.).
Flow cytometric analysis of cell apoptosis
Following incubation for 48 h, transfected cells were harvested and washed with PBS. The apoptosis rate of transfected cells was examined using an FITC Apoptosis Detection kit (Biolegend, Inc.) according to the manufacturer's protocol. Transfected cells were resuspended in 100 µl of binding buffer followed by staining with 5 µl of Annexin V–FITC and 5 µl of propidium iodide. After 15 min incubation at room temperature in the dark, stained cells were detected via flow cytometry FACScan (BD Biosciences). CellQuest version 5.1 (BD Biosciences) software was used to analyze data.
Bioinformatics analysis and luciferase reporter assay
The putative targets of miR-765 were predicted using two microRNA target prediction websites: TargetScan (http://targetscan.org/) and miRDB (http://mirdb.org/). Luciferase reporter plasmids, including pMIR-SOX6-3′-UTR wild type (wt 1 and 2) and pMIR-SOX6-3′-UTR mutant (mut 1 and 2), were chemically created by Shanghai GenePharma Co., Ltd. For the reporter assay, cells were inoculated into 24-well plates at a density of 1.0×105 cells/well 12 h prior to transfection. A miR-765 inhibitor or NC inhibitor along with the pMIR-SOX6-3′-UTR wt or pMIR-SOX6-3′-UTR mut was co-transfected into cells using the Lipofectamine 2000 reagent, based on the manufacturer's protocol. A dual-luciferase reporter assay system (Promega Corporation) was applied to measure the luciferase activity at 48 h post-transfection. The firefly luciferase activity of each well was normalized to that of the Renilla luciferase activity.
Western blot analysis
A Total Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd.) was utilized to isolate total protein from cultured cells. The concentration of total protein was determined using a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal quantities of protein were separated via 10% SDS-PAGE and transferred to PVDF (Merck KGaA). Subsequent to 2 h blocking at room temperature with 5% dried skimmed milk in TBST, membranes were incubated with primary antibodies overnight at 4°C against SOX6 (1:1,000; cat. no. ab84880) or GAPDH (1:1,000; cat. no. ab9484; each, Abcam). This was followed by further incubation with horseradish peroxidase-conjugated secondary antibodies (1:6,000; cat. no. ab6789; Abcam) for 2 h at room temperature. Protein signals were detected using an enhanced chemiluminescent reagent (GE Healthcare Life Sciences). GAPDH was used as a loading control. Quantity One software version 4.62 (BioRad Laboratories, Inc.) was utilized for the analysis of density.
Statistical analysis
SPSS 19 software package (IBM Corp.) was used for statistical analysis. Differences between groups were analyzed using a student's t-test or one-way ANOVA. A post-hoc Student-Newman-Keuls test was then utilized. A Spearman's correlation analysis was utilized to determine the correlation between miR-765 and SOX6 mRNA levels in the plasma from patients with MM. All data was presented as the mean ± standard deviation from at least three repeats of each independent experiment. P<0.05 was considered to indicate a statistical significant difference.
Results
miR-765 is upregulated in plasma from patients with MM and MM cell lines
To assess the expression status of miR-765 in MM, the expression of miR-765 was determined in the plasma cells of 27 patients with MM and 11 healthy individuals. The results of RT-qPCR revealed that miR-765 was highly expressed in plasma samples from patients with MM compared with healthy individuals (P<0.05; Fig. 1A). miR-765 expression in three human MM cell lines was then examined, including U266, MM1S and RPMI-8226. Consistently, the expression of miR-765 was higher in all three MM cell lines compared with that in the plasma samples from healthy individuals (P<0.05; Fig. 1B). These results suggest that miR-765 is upregulated in MM, and this upregulation may serve an important role in MM progression.
Figure 1.
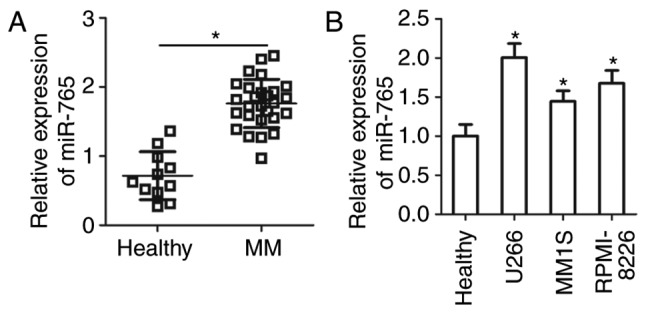
Determination of miR-765 expression in the plasma of patients with MM and human MM cell lines. (A) RT-qPCR was performed to determine miR-765 expression in the plasma of 27 patients with MM and 11 healthy individuals. (B) Expression of miR-765 in three MM cell lines (U266, MM1S and RPMI-8226) was detected via RT-qPCR. *P<0.05 vs. healthy individuals. miR-765, microRNA-765; MM, multiple myeloma; RT-q, reverse transcriptase-quantitative.
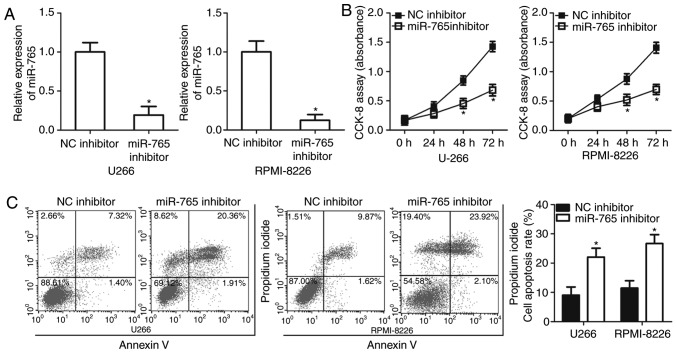
Inhibition of miR-765 attenuates cell proliferation and promotes apoptosis in MM
U266 and RPMI-8226 cell lines possessed a relatively high miR-765 expression among the three MM cell lines and thus, were selected for further functional experiments. To assess the role of miR-765 in MM cells, miR-765 was significantly knocked down in U266 and RPMI-8226 cells by transfection with a miR-765 inhibitor (P<0.05; Fig. 2A). A CCK-8 assay was then performed at different time points to determine the effect of miR-765 in MM cell proliferation. It was observed that the downregulation of miR-765 significantly impaired the proliferation of U266 and RPMI-8226 cells after 48 and 72 h (P<0.05; Fig. 2B). Since cell proliferation is closely associated with cell apoptosis, flow cytometry analysis was performed to detect the proportion of apoptotic U266 and RPMI-8226 cells after transfection with a miR-765 inhibitor or an NC inhibitor. Apoptosis rate was determined to be significantly increased in U266 and RPMI-8226 cells transfected with the miR-765 inhibitor relative to that in cells transfected with the NC inhibitor (P<0.05; Fig. 2C). The results indicate that miR-765 may serve an oncogenic role in the development of MM.
Figure 2.
miR-765 downregulation suppresses cell proliferation and induces apoptosis in MM. (A) Expression of miR-765 was evaluated in U266 and RPMI-8226 cells after transfection with miR-765 inhibitor or NC inhibitor by RT-qPCR. (B) Cellular proliferation was determined via a CCK-8 assay in U266 and RPMI-8226 cells transfected with a miR-765 inhibitor or NC inhibitor. (C) Flow cytometry analysis was performed to measure the percentage of apoptotic U266 and RPMI-8226 cells treated with a miR-765 inhibitor or NC inhibitor. *P<0.05 vs. NC inhibitor. miR-765, microRNA-765; MM, multiple myeloma; NC, negative control; RT-q, reverse transcriptase-quantitative; CCK-8, cell counting kit-8.
SOX6 is a direct target gene of miR-765 in MM cells
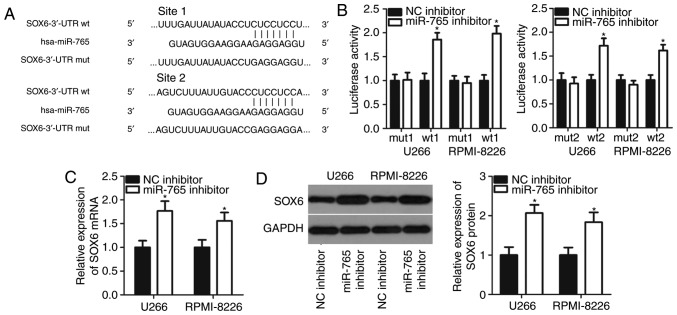
It is generally accepted that miRNAs contribute to cancer initiation and progression by regulating their target genes (6). To determine the mechanisms underlying miR-765 activity in MM cells, bioinformatics analysis was performed to search for potential targets of miR-765. Two putative miR-765-binding sites were located in the 3′-UTR of SOX6 (Fig. 3A). SOX6 was selected for further experimental validation as SOX6 is involved in the tumorigenesis and tumor development of multiple types of human cancer (28–30). A luciferase reporter assay was performed to confirm whether miR-765 directly targets the 3′-UTR of SOX6. The results revealed that co-transfection with the pMIR-SOX6-3′-UTR wt (1 and 2) and miR-765 inhibitor in U266 and RPMI-8226 cells led to a significant increase in luciferase activity (P<0.05; Fig. 3B). However, the luciferase activity in cells transfected with the pMIR-SOX6-3′-UTR mut (1 and 2) and miR-765 inhibitor was not of significance. Furthermore, the mRNA and protein levels of SOX6 were evaluated in U266 and RPMI-8226 cells upon miR-765 downregulation. The results revealed that the downregulation of miR-765 significantly increased SOX6 expression in U266 and RPMI-8226 cells at mRNA (P<0.05; Fig. 3C) and protein levels (P<0.05; Fig. 3D). These results demonstrate that SOX6 is a direct target gene of miR-765 in MM cells.
Figure 3.
SOX6 is a direct target gene of miR-765 in multiple myeloma cells. (A) Wild-type and mutant complementary sequences of miR-765 in the 3′-UTR of SOX6. (B) U266 and RPMI-8226 cells were transfected with a reporter plasmid carrying wild type or mutant binding sites along with a miR-765 inhibitor or NC inhibitor. Relative luciferase activity was detected at 48 h post-transfection. (C) Reverse transcriptase-quantitative PCR and (D) western blot analysis were performed to measure SOX6 mRNA and protein expression in U266 and RPMI-8226 cells following miR-765 inhibitor or NC inhibitor transfection. *P<0.05 vs. NC inhibitor. SOX-6, SRY-Box-6; miR-765, microRNA-765; UTR, untranslated region; NC, negative control; wt, wild type; mut, mutant.
Downregulation of SOX6 is negatively correlated with miR-765 expression in the plasma from patients with MM
To further determine the association between miR-765 and SOX6 in MM, SOX6 expression was assessed in the plasma of 27 patients with MM patients and 11 healthy individuals. RT-qPCR analysis revealed that SOX6 mRNA expression was significantly lower in the plasma of patients with MM (P<0.05; (Fig. 4A). In addition, a significant negative correlation was identified between miR-765 and SOX6 mRNA expression in the plasma of patients with MM (r=−0.5530; P=0.0028; Fig. 4B), as determined via Spearmans correlation analysis. These results suggest that the downregulation of SOX6 in MM may be, at least in part, caused by miR-765 upregulation.
Figure 4.
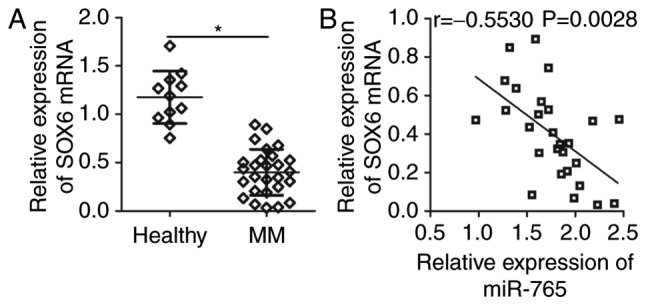
SOX6 is downregulated in the plasma of patients with MM and is negatively correlated with miR-765 levels. (A) reverse transcription-quantitative PCR detection of SOX6 mRNA expression in the plasma from 27 patients with MM and 11 healthy individuals. *P<0.05 vs. healthy individuals. (B) The correlation between miR-765 and SOX6 mRNA levels in the plasma of patients with MM was assessed through Spearman correlation analysis. r=−0.5530, P=0.0028. SOX6, SRY-Box 6; MM, multiple myeloma; miR-765, microRNA-765.
SOX6 is required for miR-765-induced phenotypes in MM cells
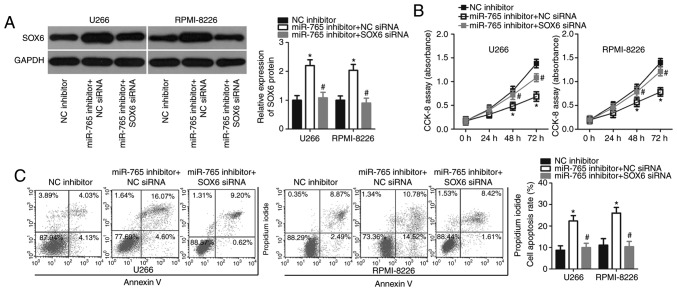
As SOX6 was validated as a direct target of miR-765, rescue experiments were subsequently performed to investigate whether SOX6 was essential for miR-765 inhibitor-induced proliferation suppression in MM cells. A miR-766 inhibitor was co-transfected with SOX6 siRNA or NC siRNA into U266 and RPMI-8226 cells. Western blot analysis revealed that the upregulation of SOX6 caused by miR-765 downregulation was recovered in U266 and RPMI-8226 cells after co-transfection with SOX6 siRNA (P<0.05; Fig. 5A). Functional assays also revealed that recovered SOX6 expression reversed the effects of miR-765 downregulation in the proliferation (P<0.05; Fig. 5B) and apoptosis (P<0.05; Fig. 5C) of U266 and RPMI-8226 cells. These data confirm that miR-765 may serve as an oncogene in MM cells, at least partly, through the regulation of SOX6 expression.
Figure 5.
SOX6 silencing abrogates the effects of miR-765 knockdown in multiple myeloma cells. The miR-765 inhibitor in combination with SOX6 siRNA or NC siRNA was co-transfected into U266 and RPMI-8226 cells. (A) The transfected cells were subjected to western blot analysis for the determination of SOX6 protein expression. The (B) proliferation and (C) apoptosis of aforementioned cells was determined via a CCK-8 assay and flow cytometry analysis, respectively. *P<0.05 vs. NC inhibitor. #P<0.05 vs. miR-765 inhibitor+NC siRNA. SOX-6, SRY-Box-6; mir-765, microRNA-765; siRNA, small interfering RNA; NC, negative control.
Discussion
Previous studies have demonstrated that miRNAs are closely associated with the occurrence and development of MM via the regulation of cell proliferation, cycle, apoptosis, angiogenesis and metastasis (31–33). miRNAs have been proposed to be novel diagnostic biomarkers and effective therapeutic targets for anticancer therapy (34). Accordingly, an in-depth understanding of the biological functions of miRNAs in MM may be helpful to identify promising therapeutic techniques for patients with MM. In the present study, a series of experiments were performed to determine the expression status of miR-765 in MM and to assess its role in MM development. The molecular mechanisms underlying the action of miR-765 in MM were also explored. The results revealed that miR-765 may serve as a diagnostic biomarker and antitumor therapeutic agent for MM.
miR-765 is aberrantly upregulated in esophageal squamous cell carcinoma tissues and cell lines (24). The upregulation of miR-765 is strongly associated with tumor stage, lymph node metastasis and clinical stage of patients with esophageal squamous cell carcinoma (24). Patients with esophageal squamous cell carcinoma and high miR-765 levels exhibit poorer overall survival and disease-free survival rates than patients with low miR-765 levels (24). Multivariate analysis has previously identified miR-765 as an independent biomarker for the prediction of overall survival and disease-free survival for patients with esophageal squamous cell carcinoma (24). miR-765 expression is also increased in osteosarcoma (25) and hepatocellular carcinoma (26). Expression levels of miR-765 are significantly correlated with the better prognosis of osteosarcoma patients (25). In contrast, miR-765 is downregulated in tongue squamous cell carcinoma (35). However, the expression pattern of miR-765 in MM remains unclear. In the current study, RT-qPCR was performed to measure the expression of miR-765 in the plasma of patients with MM and cell lines. The results revealed that miR-765 was highly expressed in the plasma from patients with MM and MM cell lines. These inconsistent results suggest a cancer-specific expression pattern of miR-765 in human malignant tumors.
miR-765 is an oncogene in hepatocellular carcinoma (26). The upregulation of miR-765 induces the proliferation and tumorigenesis of hepatocellular carcinoma cells by regulating polyphosphate 4-phosphatase type II (26). Inversely, miR-765 serves a role in tumor suppression in tongue squamous cell carcinoma. miR-765 directly targets laminin subunit gamma 2 to inhibit tongue squamous cell carcinoma cell proliferation and invasion to increase cell cycle arrest (35). However, the exact roles of miR-765 in MM remain poorly understood. To the best of our knowledge, the present study revealed that the downregulation of miR-765 restricts cell proliferation and promotes cell apoptosis in MM. These results suggest that miR-765 may serve as a novel therapeutic target for patients with these specific types of cancer.
Identifying the direct target genes of miR-765 in MM may facilitate the development of promising therapeutic targets. SOX6, a member of the Sox transcription-factor family (36), is demonstrated to be a direct target gene of miR-765 in MM cells. SOX6 is aberrantly upregulated in numerous types of cancer, including pancreatic cancer (37), prostate cancer (38), ovarian cancer (29), colorectal cancer (39) and hepatocellular carcinoma (40). SOX6 may contribute to the occurrence and development of tumorigenesis and tumor development through the regulation of various biological features, including cell proliferation, cycle, apoptosis, epithelial-mesenchymal transition and metastasis (28–30). The current study demonstrated that miR-765 directly targets SOX6 to implicate the progression of MM. The restoration of SOX6 using miR-765 based targeted therapy may therefore be considered a novel and promising therapeutic opportunity for the treatment of patients with this aggressive cancer.
In conclusion, the present study revealed that miR-765 is upregulated in the plasma of patients with MM and MM cell lines. In addition, miR-765 may serve oncogenic roles in MM by regulating cell proliferation and apoptosis in vitro. Furthermore SOX6 was identified as a direct target gene of miR-765 in MM cells. The results indicate that the inhibition of miR-765 may have great potential in inhibiting the rapid proliferation and accumulation of MM cells, confirming that miR-765 is a promising target for MM prevention and treatment. However, the effects of SOX6 in MM cells were not examined in the current study. This limitation however, should be resolved in future research.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
GC and Shifeng L designed the present study and wrote the manuscript. RT-qPCR, western blot analysis and the CCK-8 assay were performed by Shifeng L, and Shengping L. HH and GC performed flow cytometry analysis, the luciferase reporter assay and statistical analysis. All authors have read and approved the final draft of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (Nanchang, China), and was performed in accordance with the Declaration of Helsinki and the guidelines of the Ethics Committee of The First Affiliated Hospital of Nanchang University. Written informed consent was obtained from all patients for the use of their clinical tissues.
Patient consent for publication
Patient consent for publication was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- 2.Pruneri G, Cinieri S, Peccatori F, Viale G. Unusual cases in multiple myeloma and a dramatic response in metastatic lung cancer: Case 2. Plasma cell myeloma coexisting with metastatic breast carcinoma in the bone marrow. J Clin Oncol. 2005;23:232–233. doi: 10.1200/JCO.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 3.Naymagon L, Abdul-Hay M. Novel agents in the treatment of multiple myeloma: A review about the future. J Hematol Oncol. 2016;9:52. doi: 10.1186/s13045-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar SK, Lee JH, Lahuerta JJ, Morgan G, Richardson PG, Crowley J, Haessler J, Feather J, Hoering A, Moreau P, et al. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: A multicenter international myeloma working group study. Leukemia. 2012;26:149–157. doi: 10.1038/leu.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S, Hu C, Liu P, Lu M. Tumor-derived exosomes in cancer metastasis risk diagnosis and metastasis therapy. Clin Transl Oncol. 2019;21:152–159. doi: 10.1007/s12094-018-1918-0. [DOI] [PubMed] [Google Scholar]
- 8.Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Onco Targets Ther. 2018;11:3891–3900. doi: 10.2147/OTT.S156921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma N, Baruah MM. The microRNA signatures: Aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2018;21:126–144. doi: 10.1007/s12094-018-1910-8. [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, Ju S, Chu H, Shen X, Zhang Y, Luo X, Cong H. The potential function of microRNAs as biomarkers and therapeutic targets in multiple myeloma. Oncol Lett. 2018;15:6094–6106. doi: 10.3892/ol.2018.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobili L, Ronchetti D, Agnelli L, Taiana E, Vinci C, Neri A. Long Non-Coding RNAs in multiple myeloma. Genes (Basel) 2018;9(pii):E69. doi: 10.3390/genes9020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rastgoo N, Abdi J, Hou J, Chang H. Role of epigenetics-microRNA axis in drug resistance of multiple myeloma. J Hematol Oncol. 2017;10:121. doi: 10.1186/s13045-017-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang YZ, Zhang XY, Wan Q, Li J. Role of Exosomal miRNA in multiple myeloma progression and its possible mechanism-review. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25:301–305. doi: 10.7534/j.issn.1009-2137.2017.01.055. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 14.Rossi M, Tagliaferri P, Tassone P. MicroRNAs in multiple myeloma and related bone disease. Ann Transl Med. 2015;3:334. doi: 10.3978/j.issn.2305-5839.2015.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi C, Chng WJ. MicroRNA: Important player in the pathobiology of multiple myeloma. Biomed Res Int. 2014;2014:521586. doi: 10.1155/2014/521586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y, Shi X, Liu Y, Xu R, Ai Q. MicroRNA-338-3p inhibits proliferation and promotes apoptosis of multiple myeloma cells through targeting Cyclin-dependent kinase 4. Oncol Res. 2018;27:117–124. doi: 10.3727/096504018X15213031799835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang B, Xu A, Xu J, Huang H, Chen L, Su Y, Zhang L, Li J, Fan F, Deng J, et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int J Cancer. 2018;142:109–120. doi: 10.1002/ijc.31041. [DOI] [PubMed] [Google Scholar]
- 18.Alzrigat M, Jernberg-Wiklund H. The miR-125a and miR-320c are potential tumor suppressor microRNAs epigenetically silenced by the polycomb repressive complex 2 in multiple myeloma. RNA Dis. 2017;4(pii):e1529. doi: 10.14800/rd.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Chen Y, Zhao P, Zang L, Zhang Z, Wang X. MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leuk Lymphoma. 2017;58:932–940. doi: 10.1080/10428194.2016.1213827. [DOI] [PubMed] [Google Scholar]
- 20.Hua J, Ding T, Yang L. Dysfunction of microRNA-32 regulates ubiquitin ligase FBXW7 in multiple myeloma disease. Onco Targets Ther. 2016;9:6573–6579. doi: 10.2147/OTT.S105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda S, Kitadate A, Abe F, Saitoh H, Michishita Y, Hatano Y, Kawabata Y, Kitabayashi A, Teshima K, Kume M, et al. Hypoxia-inducible microRNA-210 regulates the DIMT1-IRF4 oncogenic axis in multiple myeloma. Cancer Sci. 2017;108:641–652. doi: 10.1111/cas.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Zhang G, Yu W, Gao N, Peng J. miR-186 inhibits cell proliferation in multiple myeloma by repressing Jagged1. Biochem Biophys Res Commun. 2016;469:692–697. doi: 10.1016/j.bbrc.2015.11.136. [DOI] [PubMed] [Google Scholar]
- 23.Saha MN, Abdi J, Yang Y, Chang H. MiRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced anti-myeloma activity by targeting c-Myc. Oncotarget. 2016;7:7149–7160. doi: 10.18632/oncotarget.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang B, Xu G, Lv HQ, Huang M, Li Z. Up-regulation of miR-765 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:3789–3794. doi: 10.26355/eurrev_201806_15261. [DOI] [PubMed] [Google Scholar]
- 25.Liang W, Wei X, Li Q, Dai N, Li CY, Deng Y, Jiang X, Tan XR, Dai XY, Li MX, et al. MicroRNA-765 enhances the anti-angiogenic effect of CDDP via APE1 in Osteosarcoma. J Cancer. 2017;8:1542–1551. doi: 10.7150/jca.18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie BH, He X, Hua RX, Zhang B, Tan GS, Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN, Li HP. Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomark. 2016;16:405–413. doi: 10.3233/CBM-160579. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Li J, Li K, Xu J. SOX6 is downregulated in osteosarcoma and suppresses the migration, invasion and epithelial-mesenchymal transition via TWIST1 regulation. Mol Med Rep. 2018;17:6803–6811. doi: 10.3892/mmr.2018.8681. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Xiao M, Guo F. The role of Sox6 and Netrin-1 in ovarian cancer cell growth, invasiveness, and angiogenesis. Tumour Biol. 2017;39:1010428317705508. doi: 10.1177/1010428317705508. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Xia Y, Niu H, Chen Y. MiR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1340–1348. doi: 10.1159/000358701. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Li F, Saha MN, Abdi J, Qiu L, Chang H. miR-137 and miR-197 induce apoptosis and suppress tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer Res. 2015;21:2399–2411. doi: 10.1158/1078-0432.CCR-14-1437. [DOI] [PubMed] [Google Scholar]
- 32.Qu X, Zhao M, Wu S, Yu W, Xu J, Xu J, Li J, Chen L. Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. Med Oncol. 2014;31:219. doi: 10.1007/s12032-014-0219-x. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Liu S, Wang Y. MicroRNA-21 and multiple myeloma: Small molecule and big function. Med Oncol. 2014;31:94. doi: 10.1007/s12032-014-0094-5. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Wang H, Chen E, Xu Z, Chen B, Lu G. Candidate microRNAs as biomarkers of thyroid carcinoma: A systematic review, meta-analysis, and experimental validation. Cancer Med. 2016;5:2602–2614. doi: 10.1002/cam4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47:468–476. doi: 10.1111/jop.12677. [DOI] [PubMed] [Google Scholar]
- 36.Ohta S, Misawa A, Lefebvre V, Okano H, Kawakami Y, Toda M. Sox6 up-regulation by macrophage migration inhibitory factor promotes survival and maintenance of mouse neural stem/progenitor cells. PLoS One. 2013;8:e74315. doi: 10.1371/journal.pone.0074315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Yuan Q, Jiang Y, Huang L, Chen C, Hu G, Wan R, Wang X, Yang L. Identification of Sox6 as a regulator of pancreatic cancer development. J Cell Mol Med. 2018;22:1864–1872. doi: 10.1111/jcmm.13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y, Wang Z, Sun D, Zhou X, Wei X, Hou W, Ding Y, Ma Y, Hou Y. miR-671 promotes prostate cancer cell proliferation by targeting tumor suppressor SOX6. Eur J Pharmacol. 2018;823:65–71. doi: 10.1016/j.ejphar.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Li YC, Li CF, Chen LB, Li DD, Yang L, Jin JP, Zhang B. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015;8:2981–2988. doi: 10.2147/OTT.S89459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X, Yang M, Gu H, Zhao J, Zou L. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol. 2013;37:732–736. doi: 10.1016/j.canep.2013.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.