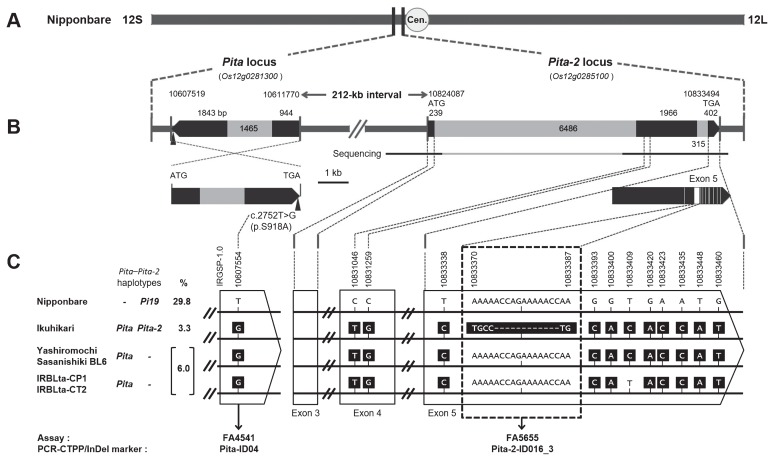
Fig. 2.
Identification of polymorphisms to discriminate Pita-2 from Pi19 and frequency of various Pita–Pita-2 haplotypes among Asian cultivated rice. (A) Positions of the Pita (Os12g0281300) and Pita-2 (Os12g0285100) loci on chromosome 12 in ‘Nipponbare’ IRGSP-1.0. 12S, short arm side of chromosome 12; 12L, long arm side of chromosome 12; Cen., centromere. (B) Gene structures of Pita and Pita-2 in ‘Nipponbare’. Black, coding regions; gray, introns. The upward arrowhead on Pita represents the position of the functional nucleotide polymorphism described previously (Bryan et al. 2000). For Pita-2, the horizontal black lines labeled “Sequencing” indicate regions where the DNA sequences of reference varieties were determined by direct sequencing, and the regions in white within exon 5 represent sites containing polymorphisms between ‘Nipponbare’ (Pi19) and ‘Ikuhikari’ (Pita-2). (C) Pita–Pita-2 haplotypes in the six cultivars, functional nucleotide polymorphisms of Pita (detected by allele-specific markers FA4541 and Pita-ID04) and polymorphisms that discriminate between Pita-2 and Pi19 (detected by allele-specific markers FA5655 and Pita-2-ID016_3). Frequency of each of the three DNA haplotypes among tested samples (Supplemental Table 1) is indicated as a percentage. The sequences within the dotted line box show variations that specifically discriminate the Pita-2 allele (‘Ikuhikari’) from the other alleles (Pi19 and susceptibility allele).