Abstract
Epistasis analysis of gid1 single and double mutants revealed that GID1c is a key positive regulator of seed germination, whereas the GID1b receptor can negatively regulate germination in dormant seeds and in the dark. The GID1 GA receptors were expected to positively regulate germination because the plant hormone gibberellin (GA) is required for seed germination in Arabidopsis thaliana. The three GA hormone receptors, GID1a, GID1b, and GID1c, positively regulate GA responses via GA/GID1‐stimulated destruction of DELLA (Asp‐Glu‐Leu‐Leu‐Ala) repressors of GA responses. The fact that the gid1abc triple mutant but not gid1 double mutants fail to germinate indicates that all three GA receptors can positively regulate non‐dormant seed germination in the light. It was known that the gid1abc triple mutant fails to lose dormancy through the dormancy breaking treatments of cold stratification (moist chilling of seeds) and dry after‐ripening (a period of dry storage). Previous work suggested that there may be some specialization of GID1 gene function during germination because GID1b mRNA expression was more highly induced by after‐ripening, whereas GID1a and GID1c mRNA levels were more highly induced by cold stratification. In light‐germinated dormant seeds, the gid1b mutation can partly rescue the germination efficiency of gid1a but not of gid1c seeds. Thus, GID1b can function as an upstream negative regulator GID1c, a positive regulator of dormant seed germination. Further experiments showed that GID1b can negatively regulate dark germination. Wild‐type Arabidopsis seeds do not germinate well in the dark. The gid1b and gid1ab double mutants germinated much more efficiently than wild type, gid1c, or gid1ac mutants in the dark. The observation that the gid1ab double mutant also shows increased dark germination suggests that GID1b, and to some extent GID1a, can act as upstream negative regulators of GID1c. Since the gid1abc triple mutant failed to germinate in the dark, it appears that GID1c is a key downstream positive regulator of dark germination. This genetic analysis indicates that the three GID1 receptors have partially specialized functions in GA signaling.
Keywords: epistasis, germination, gibberellin, GID1, seed dormancy
1. INTRODUCTION
Plant embryos can survive for extended periods of time in a dry seed, in a state resembling suspended animation (reviewed in Bewley, Bradford, Hilhorst, & Nonogaki, 2013). This allows plants to be spread by wind or animal activity, and allows humans to store crop varieties for extended periods as metabolically quiescent dry seeds. The seeds of many temperate plant species are dormant at maturity, meaning that they cannot germinate unless they experience dormancy‐breaking conditions. In Arabidopsis, seed dormancy can be released by a period of dry storage called after‐ripening or by cold stratification, taking up or imbibing water in the cold. Seed germination is a process beginning with water uptake and ending with the final germination event when the embryonic root penetrates the surrounding structures of the seed coat. Seed dormancy allows time for seed dispersal, prevents seeds from germinating out of season since a spring annual must experience an extended period of wintry cold stratification to germinate, and can help species to survive natural disasters as seeds stored in the soil.
It is well established that seed dormancy and germination are regulated by the opposing action of two plant hormones, abscisic acid (ABA) and gibberellin (GA) (reviewed by Finkelstein, Reeves, Ariizumi, & Steber, 2008). ABA is needed to establish dormancy during seed maturation, maintains dormancy in mature seeds, and can inhibit germination when externally applied to seeds. GA stimulates seed germination and is absolutely required for germination in some plant species like Arabidopsis and tomato. GA also stimulates stem elongation, the transition to flowering, and fertility. Arabidopsis seeds require light to germinate efficiently. Red light breaks Arabidopsis seed dormancy while far‐red light promotes dormancy. GA and ABA also participate in light regulation of germination. Red light stimulates GA biosynthesis and inhibits ABA biosynthesis, whereas far‐red light promotes GA turnover and stimulates ABA biosynthesis (reviewed by Yamaguchi, 2008). In Arabidopsis, the ga1 biosynthesis mutant completely fails to germinate unless GA hormone is exogenously applied or the seed coat is cut. Moreover, GA‐insensitive signaling mutants like sly1 (sleepy1) cause increased seed dormancy, whereas GA hypersensitive mutants like spy (spindly) cause decreased seed dormancy.
GA stimulates seed germination and other developmental events by lifting DELLA protein repression of GA responses (reviewed by Hauvermale, Ariizumi, & Steber, 2012; Thomas, Blázquez, & Alabadi, 2016; Urbanova & Leubner‐Metzger, 2016). DELLA proteins are named for a conserved amino acid sequence (Asp‐Glu‐Leu‐Leu‐Ala), and are a family of nuclear‐localized transcriptional regulators that negatively regulate GA signaling and responses. The partially specialized roles of the five DELLA family members in Arabidopsis have been defined by characterizing how loss of function mutations in DELLA genes suppress ga1‐3 phenotypes (Cheng et al., 2004; Dill, Jung, & Sun, 2001; King, Moritz, & Harberd, 2001; Lee et al., 2002; Tyler et al., 2004; Yu et al., 2004; Wild et al., 2012). For example, loss of the DELLA gene RGL2 rescued the germination of the GA biosynthesis mutant ga1‐3 in the light, but complete rescue of ga1‐3 dark germination required mutations in DELLA genes RGL2, RGA, and GAI. Thus, RGL2 is considered to be the main DELLA negatively regulating germination. RGA and GAI are the main DELLAs negatively regulating cell expansion, whereas RGA, RGL1, and RGL2 negatively regulate the transition to flowering. DELLA RGL3 interacts with jasmonic acid signaling to regulate plant defense responses.
GA lifts DELLA repression of seed germination and other GA responses by directing DELLA destruction via the ubiquitin‐proteasome pathway (Ariizumi & Steber, 2007; McGinnis et al., 2003; Nelson & Steber, 2016; Tyler et al., 2004). The GA hormone signal is perceived by the GIBBERELLIN‐INSENSITIVE DWARF1 (GID1) GA receptors (Nakajima et al., 2006; Ueguchi‐Tanaka et al., 2005). In the absence of GA, DELLA proteins repress seed germination and other GA responses. GA binding to the GID1 receptor results in a conformational change in the receptor, enabling it to bind DELLA protein. Formation of the GID1‐GA‐DELLA complex, causes the SLY1 F‐box protein to bind the DELLA protein (Ariizumi, Lawrence, & Steber, 2011; Wang & Deng, 2011). SLY1 is the F‐box subunit of an SCF E3 ubiquitin ligase that catalyzes DELLA polyubiquitination, causing DELLA to be degraded by the 26S proteasome. DELLA destruction allows seeds to germinate.
Since GID1 and SLY1 are both positive regulators of GA signaling, loss of function should result in increased seed dormancy/reduced germination. The sly1 mutants are GA‐insensitive dwarves that exhibit increased seed dormancy (Ariizumi & Steber, 2007). The sly1‐2 allele requires one to two years to after‐ripen, whereas the stronger sly1‐t2 fails to after‐ripen. Rescue of sly1‐2 germination by after‐ripening and by overexpression of the GID1 receptor genes on the CaMV 35S promoter is associated not with decreased DELLA protein accumulation, but with increased formation of the GID1‐DELLA protein complex (Ariizumi et al., 2013; Fukazawa, Ito, Kamiya, Yamaguchi, & Takahashi, 2015). It appears that GID1‐binding to DELLA blocks DELLA interaction with downstream regulators of GA signaling.
The GID1a, GID1b, and GID1c genes appear to function redundantly as positive regulators of seed germination in the light (Griffiths et al., 2006; Iuchi et al., 2007; Voegele, Linkies, Mueller, & Leubner‐Metzger, 2011; Willige et al., 2007). The fact that the gid1a gid1b gid1c triple mutant is unable to germinate unless the seed coat is cut, while the gid1 single mutants can germinate suggests that the three genes act redundantly to stimulate germination. Moreover, a positive role in seed germination is suggested by the fact that overexpression of each Arabidopsis GID1 gene increases sly1 mutant germination and increases the GA‐sensitivity of ga1‐3 during germination (Ariizumi et al., 2013; Hauvermale, Ariizumi, & Steber, 2014; Hauvermale, Tuttle, Takebayashi, Seo, & Steber, 2015). Interestingly, overexpression of the GID1b gene more strongly stimulated germination than overexpression of the GID1a or GID1c genes. This suggested that there might be some specialization in GID1 gene function in seed germination.
In order to examine whether there may be more specialization in GID1 gene function during seed germination, this study closely examined the relative effects of gid1 single and double mutants on initial Arabidopsis seed dormancy, dormancy loss, and germination. We reasoned that conditions likely to inhibit germination would accentuate the germination phenotype of gid1 single and double mutants allowing us to better examine their relative effects on seed dormancy and germination. Voegele et al. (2011) provided a careful examination of the germination phenotypes of after‐ripened gid1 mutants germinated in the light. Using the same gid1 alleles, this study examined the effect of gid1 single and double mutants on initial dormancy, after‐ripening, dark germination, and GA sensitivity. Examination of mutant phenotypes told a new story about GID1 regulation of germination. GID1b appeared to behave like a negative regulator of germination in dormant seeds germinated in the light and in dark‐germinated seeds. GID1c is a strong positive regulator of germination, whereas the role of GID1a appears to depend upon dormancy and lighting conditions.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
The gid1a‐1, gid1b‐1, and gid1c‐2 single mutants, double mutants, and segregating triple mutant (gid1b/gid1b gid1c/gid1c gid1a/+) in the Arabidopsis thaliana ecotype Columbia (Col‐0) background were a gift from Claus Schwechheimer (Willige et al., 2007). Each of these three alleles contains a T‐DNA insertion in the major second exon of the GID1 gene, predicted to result in a loss‐of‐function. All genotypes were confirmed by PCR using Taq (NEB) and the primers reported by Willige et al., 2007. Genomic DNA was prepared according to McKinney et al., 1995, and PCR was performed using the PCR program: 3 min at 95°C, 42 cycles of 1 min at 95°C, 50 s at the annealing temperature, 1 min at 72°C, then 10 min at 72°C. To make sure that seeds used were exposed to the same environment during development and were of similar age, all gid1 mutants and the Columbia (Col‐0) wild type (WT) were grown side‐by‐side in a Conviron growth chamber under a 16 hr day under fluorescent lamps at 200 μmol m−2 sec−1 at 22°C. The accession numbers for GID1a, GID1b, and GID1c are AT3G05120, AT3G63010, and AT5G27320, respectively.
In order to obtain high initial seed dormancy, seeds were harvested from plants near physiological maturity, when 40%–60% of the siliques were yellow but some were still green (as in Nelson, Ariizumi, & Steber, 2017). When such plants were hand threshed, only brown mature seeds, not green seeds, were obtained. Experiments were performed using two independent sets of seeds, and the second set of seeds had more initial dormancy than the first. For dry after‐ripening (AR), seeds were stored in open tubes at room temperature (~22°C) and 15%–20% humidity for 2 days (d), 1 week (wk), 2 weeks, or 4 weeks. Light germination experiments were plated immediately upon reaching the desired after‐ripening time point. The dark germination plating experiments were performed using seed stored at −20°C for less than 2 months to maintain dormancy.
2.2. Light germination
For all germination experiments, 80–120 seeds were sterilized with 10% (v/v) bleach and 0.2% (v/v) Triton X‐100 for 10 min, and washed six times with sterile deionized water. Seeds were plated on MS‐agar plates containing 0.8% (w/v) agar and 0.5× Murishige and Skoog salts (MS, Sigma‐Aldrich), and germination was scored daily for 7 days (d) during incubation under fluorescent light (60 μmol m−2 s−1) at 22°C. For the GA dose–response experiments, 2 and 4 weeks (wk) after‐ripened seeds were incubated with gentle agitation in 10 μM paclobutrazol (PAC) in 0.5× MS salts and 5 mM MES (2‐(N‐morpholino) ethanesulfonic acid), pH 5.5; Sigma) for 48 hr in dark at 4°C to suppress endogenous GA biosynthesis, then washed 6 times with sterile water before plating on MS‐agar containing 0 μM, 0.01 μM, 0.1 μM and 0.5 μM gibberellin A3 (GA3, Phytotechnology) Germination was scored daily for 5 days and averaged over 4 biological replicates.
2.3. Dark germination
For dark germination experiments, 2 weeks and 4 weeks after‐ripened (AR) seeds were sterilized under 0.73 μmol m−2 s−1 light followed by plating under multiple fluorescent light intensities of: (a) Bright light of 16.9 μmol m−2 s−1, (b) Dim light of 1.3 μmol m−2s−1, and (c) Dimmer light of 0.3 μmol m−2 s−1. Cool white 40W fluorescent lamps were used to generate varying light intensities (Sylvania FO30/841/XP/SS/ECO3 and FO40/741/ECO). Seeds were also plated under a green LED lamp (Plant Safe 9‐LED Mini Flashlight, Hydroponics Nation) with a light intensity of 1.0 μmol m−2 s−1. Plates were wrapped with two layers of aluminum foil (one silver sheet and one black sheet), then incubated at 22°C for 14 days in the dark. Germination was scored only once under a dissection microscope. The average germination rate was calculated using at least three independent replicates. An ANOVA was performed in SPSS v19.0 to determine which germination differences were significant as shown using letters for statistical classes in Supporting Information Figure S2.
2.4. Determining seed size
Seed size was determined using two independent sets of gid1 single and double mutant, and of Col WT growth side‐by‐side in two greenhouse experiments. For each set, seeds of each genotype were pooled. We stochastically selected three subsamples of 500 seeds from each set of the pooled samples. Seeds were counted under a dissection microscope (Leica), and then the mass of each subsample determined using a quantitative balance (Denver Instruments).
3. RESULTS
3.1. Effects of gid1 single and double mutants on light germination
The effect of gid1 single and double mutants on dormancy and after‐ripening were examined by comparing light germination to Col WT at 2 days, 1, 2, and 4 weeks of dry after‐ripening. The gid1a and gid1c, but not gid1b, single mutants showed higher initial dormancy than WT (Figure 1a). Of the double mutants, gid1ac had the strongest seed dormancy, showing a complete failure to germinate through 2 weeks after‐ripening (Figure 1b). All mutants showed increasing germination with after‐ripening (Figure 1a–h). The gid1ac double mutant reached 41% germination by 4 weeks after‐ripening. To determine if gid1ac seeds were viable, seeds coats were cut at 2 weeks‐AR resulting in 86% germination within 5 days of imbibition. Since embryos are sometimes injured when the seed coat is cut, this suggests that the gid1ac seeds are dormant but viable. This is consistent with previous work showing that the gid1ac double mutant seeds are viable based on tetrazolium staining (Voegele et al., 2011). These results suggest that GID1a and GID1c are the main GA receptors serving as positive regulators of germination in the light. Previous work showed that the gid1a gid1b gid1c triple mutant fails to germinate and does not after‐ripen (Willige et al., 2007). However, no single or double gid1 mutant is sufficient to prevent after‐ripening, suggesting that the three GID1 genes act additively to positively regulate seed germination and dormancy loss.
Figure 1.
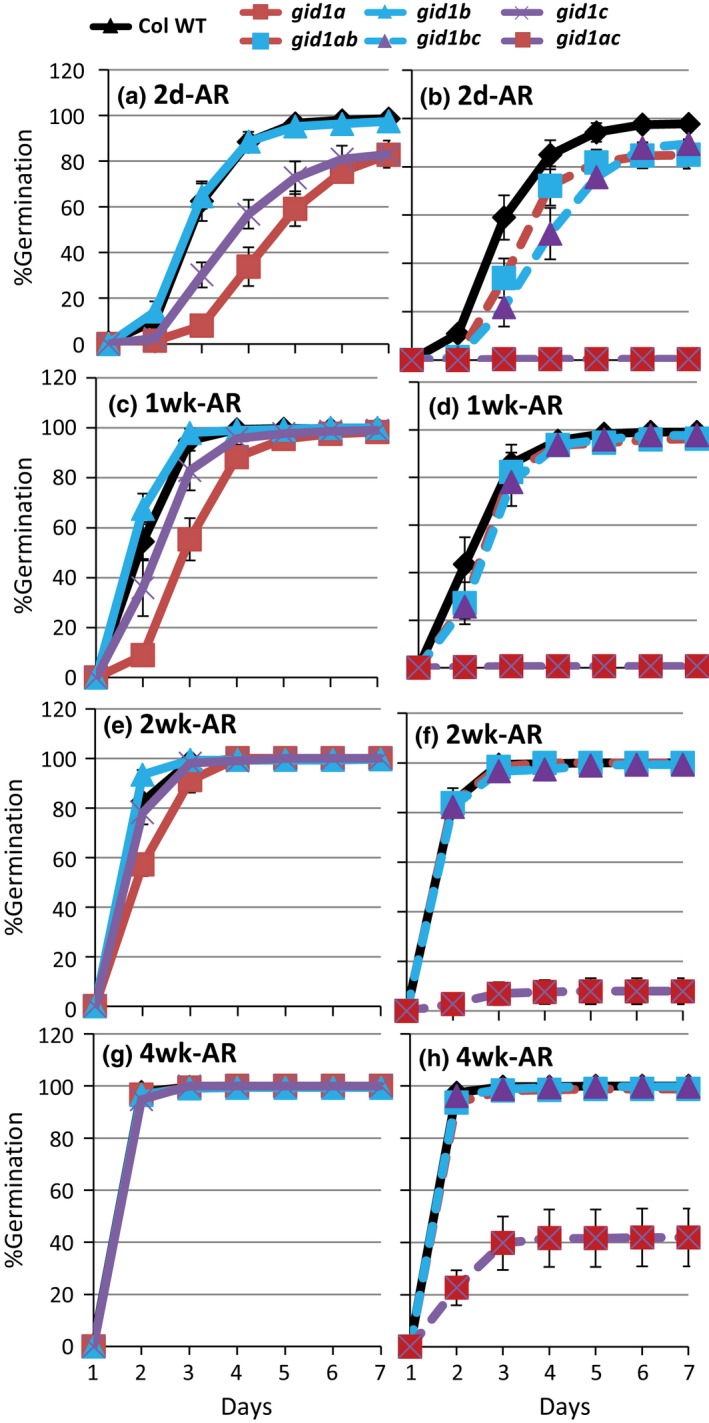
After‐ripening time course of gid1 germination. An after‐ripening time course compared gid1 single (a,c,e,g) and double (b,d,f,h) mutant seed germination. Percentage germination was scored daily over 7 days imbibition on 0.5× MS‐agar at 22°C, over four dry after‐ripening time points (1 days, 1 week, 2 weeks and 4 weeks). Mean germination is shown with error = SD, n = 3
Another story about the relative roles of GID1a, GID1b, and GID1c emerged from an epistasis analysis comparing single mutant parents to corresponding double mutant germination at 2 days after‐ripening. Consistent with the notion that GID1a and GID1c both positively regulate seed germination, the gid1a and gid1c mutations had an additive effect on seed germination. The gid1a gid1c double mutant failed to germinate, whereas the gid1a mutant showed 33.9% and that gid1c mutant 56.8% germination with 4 days incubation (Figure 2a). If GID1b were a weak positive regulator of seed germination, then you would expect the gid1bc and gid1ab germination to be similar to or reduced compared to the gid1c and gid1a parents. The gid1bc double mutant germination (52.3%) was similar to gid1c (56.8%), but lower than gid1b germination (88.5%) at 4 days incubation (Figure 2c). Thus, gid1c is epistatic to gid1b. In contrast, the gid1ab double mutant had a germination phenotype that was intermediate (72.1%) between gid1a (33.9%) and gid1b (88.5%) at 4 days incubation (Figure 2b). Thus, the gid1b mutation appears to partly rescue gid1a germination, suggesting that GID1b acts as a negative regulator of germination in dormant seeds. The observation that gid1b and gid1a have additive effects suggests that they act in parallel. The fact that gid1b can partly rescue gid1a but not gid1c germination suggests that GID1b functions upstream of GID1c as a negative regulator of seed germination. The gid1ab germination appears depend on stimulation of germination by GID1c.
Figure 2.
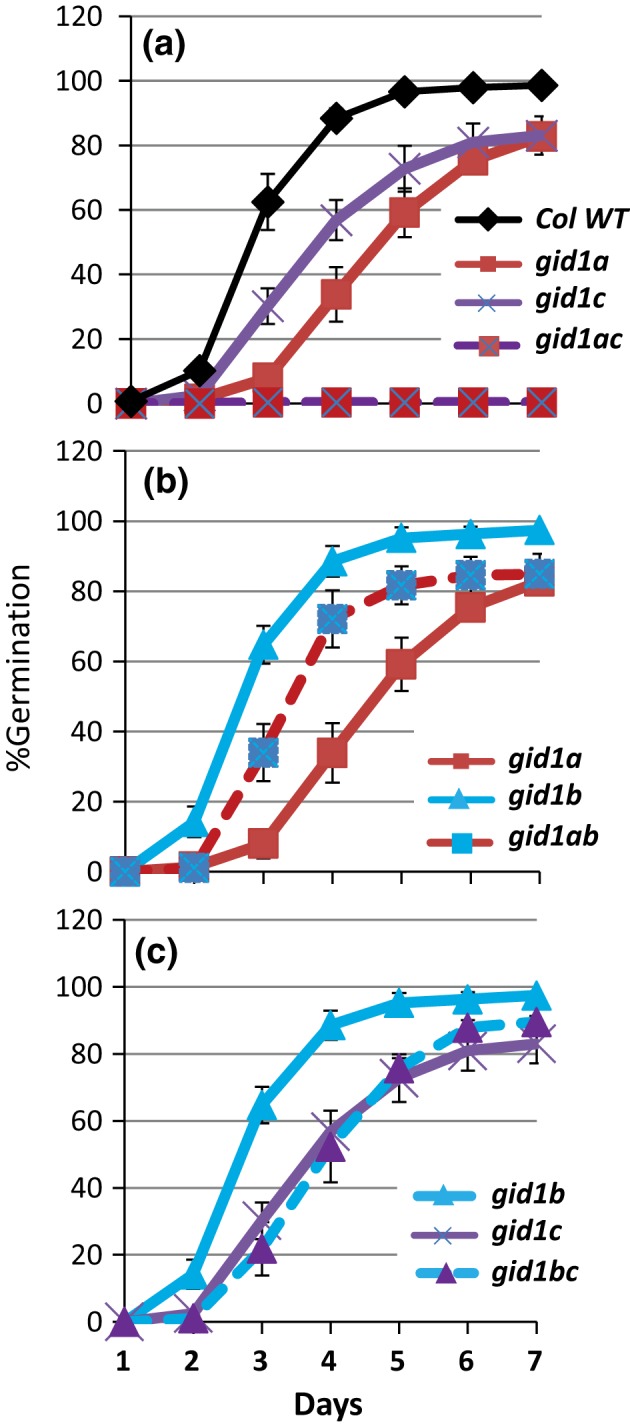
Epistasis analysis of dormant gid1 seed germination in the light. Compared are: (a) gid1a and gid1c to gid1ac, (b) gid1a and gid1b to gid1ab, and (c) gid1a and gid1c to gid1ac. Seeds were germinated at 2 days AR (as in Figure 1a,b). Error = SD, n = 3
3.2. The effect of gid1 single and double mutants on GA dose–response during light germination
GID1 GA receptor mutants were expected to have decreased ability to respond to GA during seed germination. The relative effects of the gid1 single and double mutants on GA dose–response curves were examined. The GA biosynthesis inhibitor paclobutrazol (PAC) was used to suppress endogenous GA biosynthesis in WT and gid1 mutant seeds before plating on increasing concentrations of GA3 (Supporting Information Figure S1 ABCD). The gid1b mutant showed a very similar GA dose–response to Col WT. The gid1a and gid1c single mutants and the gid1ac double mutant showed the strongest decrease in GA dose–response. The relative GA dose–response curves of the gid1 single and double mutants were similar at 2 and 4 weeks of after‐ripening.
An epistasis analysis was conducted comparing gid1 double mutant GA sensitivity to single mutant parents at 4 weeks after‐ripening (Figure 3). The gid1a and gid1c mutations had an additive effect, such that the gid1ac double mutant was less GA sensitive than the parents (Figure 3a). The gid1bc double mutant did not differ significantly from the gid1c mutant which was less GA sensitive than the gid1b mutant (Figure 3c). The gid1ab double mutant phenotype varied with GA concentration, appearing similar to gid1a at 0 μM GA, intermediate between gid1a and gid1b at 0.01 μM, and similar to gid1b at 0.1 and 0.5 μM GA (Figure 3b).
Figure 3.
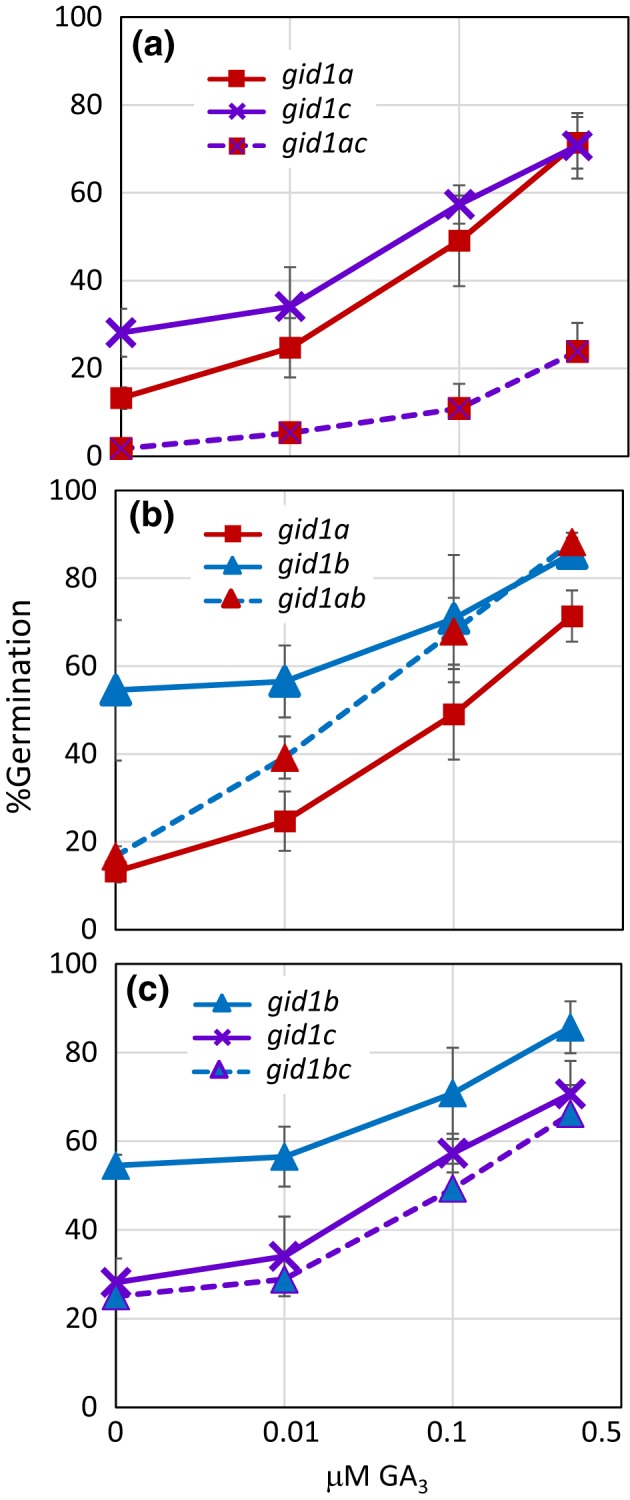
Epistasis analysis of gid1 mutant GA dose–response. Compared are light germination of: (a) gid1a and gid1c to gid1ac, (b) gid1a and gid1b to gid1ab, and (c) gid1a and gid1c to gid1ac. Seeds were incubated in 10 μM PAC for 48 hr at 4°C in dark, washed, and then incubated in the light for 5 days at 22°C on MS‐agar containing 0, 0.01, 0.1 and 0.5 μM GA 3. Germination was scored daily. The x‐axis is log10 scale. Error = SD (n = 4) is shown
3.3. The effect of gid1 single and double mutants on dark germination
We expected germination in the dark to exacerbate the gid1 seed dormancy phenotypes, making it easier to compare the relative effects of gid1 single and double mutants on germination capacity. Experiments were conducted at 2 and 4 weeks of after‐ripening using seeds that had been plated under varying light conditions referred to as: bright (16.9 μmol m2 s−1), dim (1.3 μmol m−2 s−1), and dimmer (0.3 μmol m−2 s−1) fluorescent light, and green light (1.0 μmol m−2s−1). Seed germination was scored after dark incubation for 14 days at 22°C. While similar trends were observed at 2 and 4 weeks of after‐ripening, the 4 weeks after‐ripening data are described in more detail because it is easier to observe significant differences when the germination rates are higher (Supporting Information Figure S2AB; Supporting Information Table S1; Supporting Information Table S2). The effects of gid1 mutants on dark germination varied with light intensity during plating. Generally, higher fluorescent light intensity during plating stimulated dark germination in WT and in all gid1 mutants, and green light resulted in the least efficient seed germination (Supporting Information Figure S3).
Many of the effects of the gid1 single and double mutants were observed regardless of lighting during plating, although some effects were clearer under one or more condition. Of the single mutations, only gid1c showed a significant decrease in dark germination compared to WT at 4 weeks AR and only after plating under bright lighting (Figure 4; Supporting Information Figure S2). Surprisingly, gid1b consistently germinated more efficiently than Col WT in the dark. The gid1a mutation did not show a significant decrease in germination compared to Col WT, and actually germinated more efficiently than WT when 4 weeks AR seeds were plated under green light. Thus, GID1c behaves like a strong positive regulator and GID1b a negative regulator, while GID1a behaves as a weak positive or negative regulator of dark germination depending on conditions.
Figure 4.
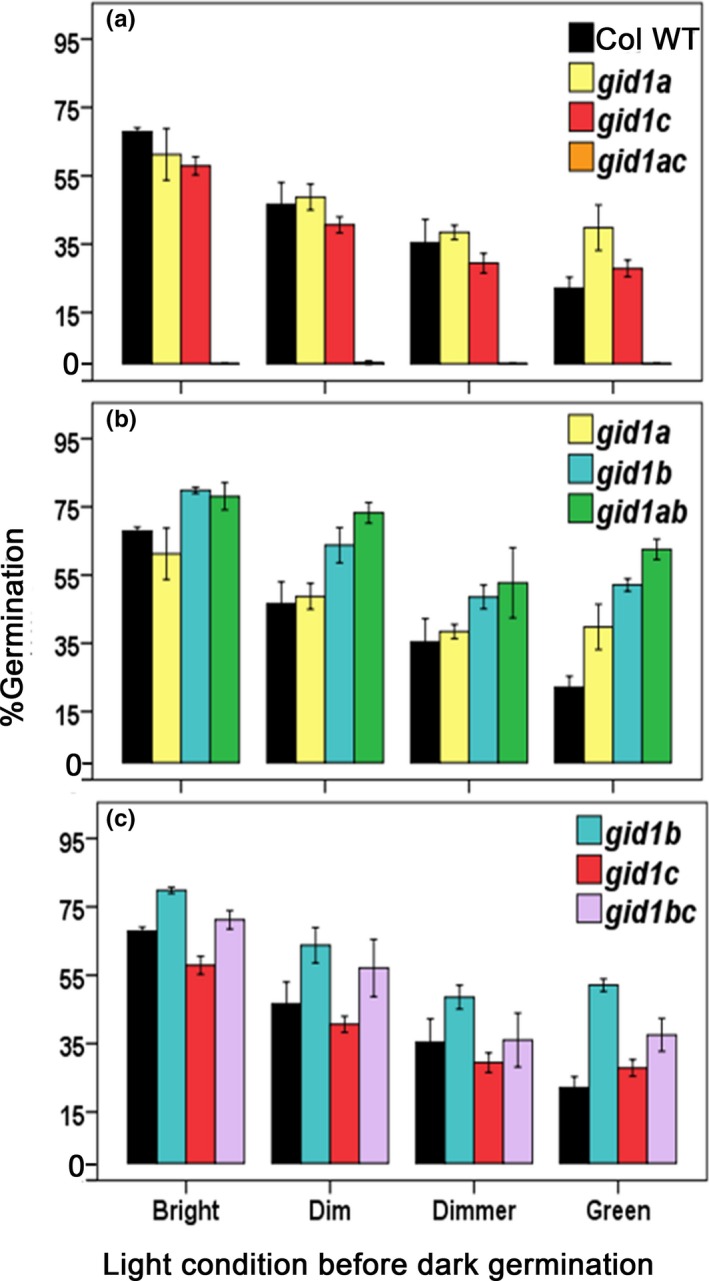
Epistasis analysis of the gid1 dark germination phenotype. Compared in order are dark germination of: (a) gid1a and gid1c to gid1ac, (b) gid1a and gid1b to gid1ab, and (c) gid1a and gid1c to gid1ac. Seeds were plated at 4 weeks AR. The x‐axis labels “Bright”, “Dim”, “Dimmer” indicate the fluorescent light intensity or quality during plating prior to incubation for 14 days in the dark at 22°C. Shown is the mean ±SE (n = 3)
An epistasis analysis of dark germination was made by comparing double mutants to single mutant parents (Figure 4). The experiment was repeated using a second batch of seeds that had higher initial dormancy (Supporting Information Figure S4). The gid1 mutants showed similar effects and epistasis relationships in the two experiments. The gid1ac double mutant failed to germinate in the dark and, therefore, germinated less efficiently than either gid1a or gid1c mutants (Figure 4; Supporting Information Figure S4). This additive effect suggests that GID1c and GID1a act in parallel as positive regulators of dark germination. Interestingly, the gid1ab double mutant germinated more efficiently than either the gid1a or gid1b single mutants when plated under green or dim lighting conditions, suggesting that GID1a can also act as a negative regulator of dark germination. Because the second set of seeds had more initial dormancy, it was more apparent that the gid1a mutation mildly stimulated germination, and that the gid1ab double mutant germinated more efficiently than the gid1b single mutant. Thus, GID1a is a weak negative regulator of dark germination in the presence of GID1c. Thus, GID1c is a strong positive regulator of seed germination that appears to be negatively regulated by GID1b and GID1a. The role of GID1a appears to be context dependent, acting as a positive regulator when GID1b is present and acting as a negative regulator when GID1c is present. These results strongly suggest that the Arabidopsis GA receptors have specialized roles in dark seed germination. The gid1bc double mutant germinated more efficiently than gid1c but less efficiently than gid1b. This suggests that GID1c and GID1b act in parallel to positively and negatively regulate dark germination, respectively. If GID1b and GID1c do act in parallel, then we might expect the gid1abc triple to germinate in the dark in a manner similar to gid1ab.
Previous work showed that the gid1abc triple mutant fails to germinate in the light unless germination is rescued by cutting the seed coat (Willige et al., 2007). However, the effect of the gid1abc triple mutation on dark germination is unknown. Since the gid1abc triple homozygous mutant is infertile, seeds of the gid1 b/b c/c a/+ line that segregates for the gid1abc triple homozygous mutant were plated to compare dark and light germination phenotypes (Figure 5). Seeds of wild‐type Col, gid1bc, gid1ab, and gid1ac were included for comparison. Seeds were sterilized and plated under fluorescent (17 μmol m−2 s−1) lights and then incubated at 22°C either under lights or in the dark. The gid1b/b c/c a/+ line should segregate 25% gid1abc triple homozygous mutant, 50% gid1b/b c/c a/+ heterozygotes, and 25% gid1b/b c/c double homozygous mutant. This line was expected to show about 75% germination under lights, and actually showed 70% germination. In the dark, the segregating line showed 0% germination, Col WT and gid1bc about 2% germination, gid1ac 0% germination, and gid1ab 39% germination. If the gid1abc triple germinated as well as the gid1ab double, then the segregating line should show about 10% germination (25% × 39%). Thus, the dark germination of the gid1abc triple does not resemble the gid1ab double mutant, indicating that GID1c must be present for gid1b and gid1a mutations to rescue dark seed germination. Thus, GID1c appears to act downstream of GID1b and GID1a to stimulate seed germination, and GID1b and GID1a appear to negatively regulate germination via GID1c.
Figure 5.
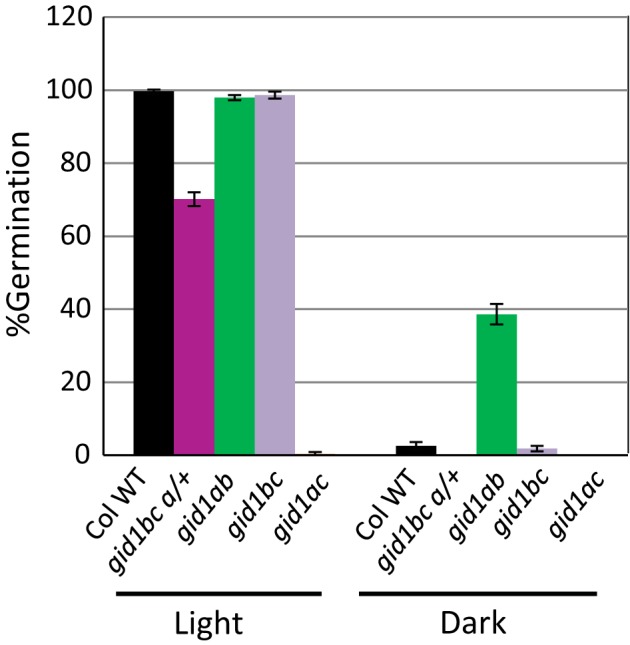
Light and dark germination of the gid1abc segregating line. Seeds of Col WT, gid1ab, gid1ac, gid1bc, and of the gid1 b/b c/c a/+ line segregating for gid1a, were plated at 4 weeks‐AR under fluorescent lights (17 μmol m−2 s−1), then germinated either in the dark or under lights at 22°C. Error bars indicate SD (n = 3)
3.4. The effect of gid1 single and double mutants on seed size
While performing germination experiments, we observed some morphological differences in the gid1 mutant seeds themselves. For example, the gid1ab seeds appeared to be larger than Col WT, whereas the gid1ac double mutant seeds appeared somewhat shriveled (Figure 6a,b). To examine whether there was a quantitative difference in seed size, we measured the dry weight of 500 seeds of Col WT, the gid1 single, and gid1 double mutants in the two sets of seeds harvested at different times (Figure 6c,d). The gid1ab double mutant seeds were significantly larger and the gid1ac double mutant, significantly smaller than WT seeds in both experiments (p < 0.001). Thus, GID1 genes appear to regulate Arabidopsis seed size, suggesting that they play a role during seed development as well as during seed germination.
Figure 6.
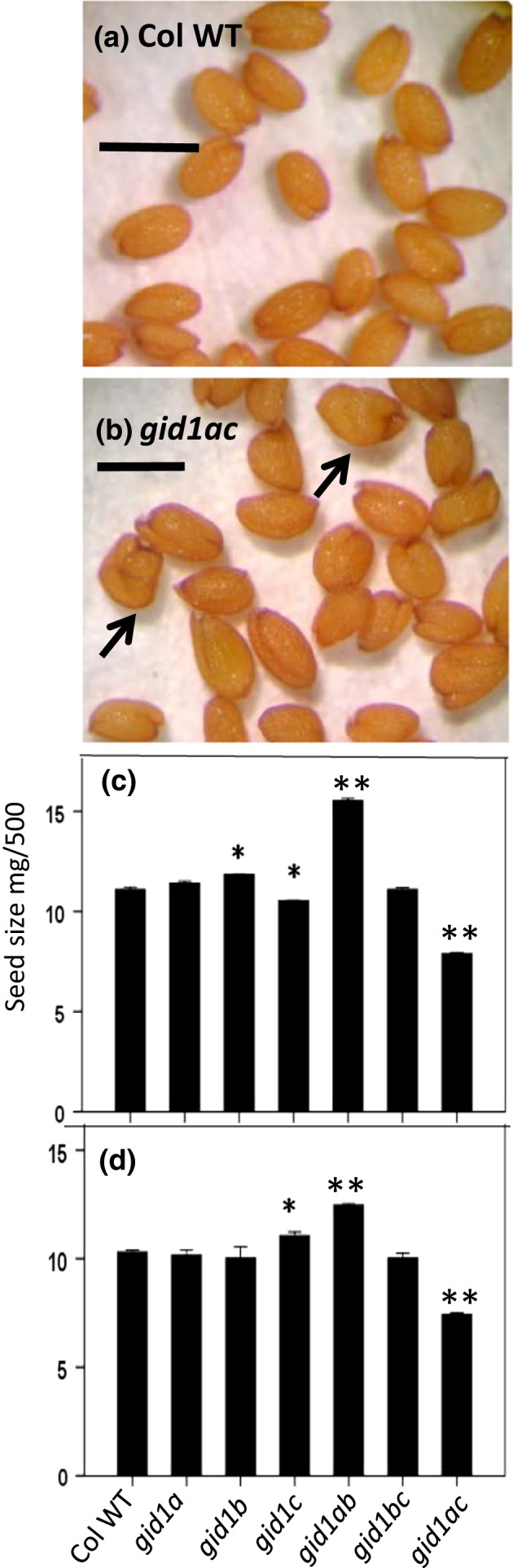
Effect of gid1 genotype on seed size. Comparison of: (a) Col WT and (b) gid1ac seed morphology showing that gid1ac seeds appear shriveled. Seed size was evaluated in two independent sets of seeds, (c) Set 1 and (d) Set 2. Student's t test was used to examine the statistical significance of seed size differences from WT, *p < 0.05, **p < 0.001. The bar indicates 0.5 mm
4. DISCUSSION
GA hormone stimulates germination in many plant species, and it is known that GA hormone biosynthesis is required for germination in Arabidopsis and tomato (reviewed in Finkelstein et al., 2008). Based on this fact, positive regulators of GA signaling like the F‐box protein SLY1 and the GID1 GA receptors are expected to function as positive regulators of seed germination. The Arabidopsis GID1a, GID1b, and GID1c genes can all function as positive regulators of seed germination because the GA‐insensitive gid1a gid1b gid1c triple mutant cannot germinate and fails to after‐ripen (Willige et al., 2007). Epistasis analysis of germination under multiple conditions, however, revealed that some of the GID1 receptors can behave as negative regulators of seed germination depending upon the genetic, developmental, and environmental context. GID1c functions as a positive regulator of seed germination in both the light and in the dark. GID1a functions as a positive regulator of seed germination in the light, but can behave like a mild negative regulator of GID1c and seed germination in the dark. GID1b appears to function as a negative regulator of seed germination in the dark and in dormant seeds germinating in the light when GID1c is present (Figure 7).
Figure 7.
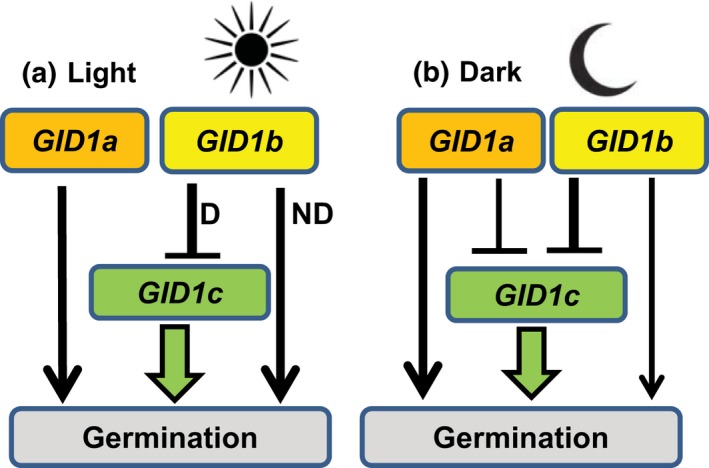
Model for GID1 regulation of seed germination. (a) In the light, GID1c and GID1a act in parallel as positive regulators of seed germination. GID1b acts as a positive regulator of seed germination in nondormant (ND) seeds, but as a negative regulator of GID1c and seed germination in dormant (D) seeds. (b) In the dark, GID1c is the stronger and GID1a the weaker positive regulator of germination. Both GID1b and GID1a can negatively regulate germination via negative regulation of GID1c
The gid1ab and gid1ac double mutants also reproducibly impacted seed size and morphology. This is interesting given that previous work suggested a role for GID1 in control of ovule development (Ferreira, et al., 2008). The gid1ab double mutant produced larger seeds than WT (Figure 6). Larger seed size is sometimes associated with higher germination efficiency, but it is more often associated with better seedling emergence and vigor, presumably due to the presence of more stored reserves (Chaisurisri, Edwards, & El‐Kassaby, 1992; Edwards & Hartwig, 1971; Gómez, 2004; Leishman, Wright, Moles, & Westoby, 2000; Wu & Du, 2007). Increased seed size alone is not sufficient to explain the increased dark germination, since the gid1a and gid1b single mutants did not consistently increase seed size but did increase in dark germination compared to WT (Figure 4; Figure 6). It is possible, however, that the larger gid1ab double mutant seeds better supported seedling growth in the dark by providing more stored reserves to compensate for lack of photosynthesis. The gid1ac double mutant seed appears to be viable based on the fact that germination was rescued by nicking the seed coat and based on tetrazolium staining by Voegele et al., 2011;. However, gid1ac mutants produced smaller, shriveled seeds (Figure 6). The shriveled phenotype of the gid1ac double mutant, while not as extreme, was reminiscent of the shriveled seed phenotype observed in the ZHOUPI (ZOU) and INDUCER OF CBF EXPRESSION 1 (ICE1) bHLH transcription factor mutants (Denay et al., 2014). The zou and ice1 mutants fail to degrade the endosperm during seed development, leading to a decrease in embryo expansion. The gid1ac embryos appeared to have developed to a normal shape and size when the seed coat was cut to stimulate germination. Future work will need to examine if gid1ac causes altered endosperm development.
Previous studies of loss‐of‐function alleles indicated that GID1a, GID1b, and GID1c act additively as positive regulators of seed germination in the light, but suggested that one GID1 gene or another may play a more prominent role (Iuchi et al., 2007; Voegele et al., 2011; Willige et al., 2007). This study examined the same gid1 mutant alleles used in both the Willige et al., 2007 and Voegele et al., 2011 studies. In the Voegele et al., 2011 study, the after‐ripened gid1c single mutant had a slightly lower, and gid1b a slightly higher, germination rate than Col WT. But those differences were not significant. Similarly, we found that none of the gid1 single mutants had a statistically significant effect on germination when after‐ripened for 4 weeks (Figure 1). In seeds showing mild dormancy (2 day after‐ripened), however, the gid1b mutation was able to improve gid1a germination efficiency, but not gid1c germination in the light (Figure 2). Thus, loss of GID1b as a negative regulator of dormant seed germination can stimulate germination when GID1c is present, suggesting that GID1b may sometimes function as a negative regulator of the downstream positive regulator GID1c (Figure 7). The notion that GID1b can negatively regulate germination is supported by the fact that the gid1b and the gid1ab double mutants showed a strong increase in dark seed germination compared to Col WT (Figure 4; Supporting Information Figure S4). The gid1a mutation only showed increased dark germination compared to WT under green light or in more dormant seeds. This suggests that GID1b, and to a lesser extent GID1a, can act as negative regulators of dark seed germination. GID1c appears to be a key positive regulator of seed germination. This is interesting given that of the gid1 single mutants, gid1c had the strongest decrease in GA dose–response (Figure 3).
These results were the reverse of what we initially expected based on overexpression experiments (Ariizumi et al., 2013; Hauvermale et al., 2014, 2015). GID1b overexpression (GID1b‐OE) better rescued the germination of the GA‐insensitive sly1‐2 mutant than overexpression of GID1c and GID1a. GID1b‐OE also better enhanced the GA sensitivity of the GA biosynthesis mutant ga1‐3. It may be that expression on the CaMV 35S promoter does not result in GID1b expression at the appropriate time or place for it to function as a negative regulator. We do know that 35S:HA:GID1a, 35S:HA:GID1b, and 35S:HA:GID1c protein expression cannot be detected at all in dormant seeds but is detected in after‐ripened seeds (Hauvermale et al., 2015). An antibody that detected all three GID1 proteins detected low level GID1 protein expression in dormant seeds that increased with after‐ripening. Thus, we suspect that HA:GID1b protein may not be present to block germination of dormant seeds when expressed on the 35S promoter. A second possibility is that GID1b cannot function as a negative regulator of germination in the absence of GA and SLY1. Future work will need to examine whether GID1 protein function in germination is subject to complex post‐translational regulation. These conflicting results point out the importance of interpreting overexpression results with caution. Another caveat is that the ga1‐3 and sly1‐2 mutants are in the Landsberg erecta (Ler) ecotype whereas the gid1 mutants are in the Col ecotype. Future work will need to examine the effects of GID1 gene overexpression in the Col ecotype. Ecotype can impact the phenotypes of GA receptor mutants given that the gid1b‐1 mutation in the Columbia‐0 ecotype showed little or no reduction in GA sensitivity, whereas another gid1b allele in the Nossen ecotype showed a strong reduction in GA sensitivity (Figure 3; Iuchi et al., 2007).
Other lines of evidence raised the possibility that the Arabidopsis GA receptors have evolved specialized functions. Based on predicted amino acid sequence alignment, Arabidopsis GID1a more closely resembles GID1c and the sole copy of GID1 in rice (Oryza sativa, OsGID1) than GID1b (Nakajima et al., 2006; Yamamoto et al., 2010). GID1a has 85% amino acid identity with GID1c, but only 66% amino acid identity with GID1b (Nelson & Steber, 2016). This difference in amino acid similarity is associated with functional differences in DELLA and GA‐binding activities (Nakajima et al., 2006; Yamamoto et al., 2010). Rice GID1 and GID1a/c‐type receptors absolutely require GA in order to bind DELLA protein, whereas GID1b‐type receptors have a limited ability to bind DELLA without GA that is enhanced by GA hormone. A single amino acid change in the rice OsGID1a (P99A) converted this receptor to a GID1b‐like, GA‐enhanced and GA‐hypersensitive receptor. The GA‐enhanced GA signaling suggests that GID1b is fundamentally different from GID1a and GID1c. The three GA receptors also differ in their affinity for GA hormone. GA associated with GDI1b more quickly than GID1a and GID1c, while GA dissociated from GID1a more slowly than from GID1c and GID1b. It is possible that the stronger rescue of sly1‐2 germination by GID1b‐OE is due to either to a lower GA requirement or to higher DELLA protein affinity. The GID1 transcripts also show differences in mRNA expression during germination (Hauvermale et al., 2015). GID1a and c transcripts are more strongly induced by cold stratification than GID1b, whereas GID1b transcript showed higher relative expression levels in the dark. These expression differences initially prompted us to more closely examine the relative function of GID1 genes during germination.
Future work will need to examine whether GID1 genes have specialized functions in other plant species or in other GA responses. While monocots tend to have a single GID1a/c‐type receptor like rice, sequence analysis showed that multiple eudicot species have both GID1a/c‐type and GID1b‐type GA receptors such as soybean (Glycine max), garden cress (Lepidium sativum), tomato (Solanum lycopersicum), alfalfa (Medicago truncatula), and oilseed rape (Brassica napus) (Voegele et al., 2011; Yamamoto et al., 2010). Functional analysis is needed to examine if GID1b can act as a negative regulator of seed germination in these plant species. Future work should also explore whether GID1b can negatively regulate other GA‐stimulated responses such as cell expansion, transition to flowering, and fertility. GID1a and GID1c are important positive regulators of stem elongation since the gid1ac double mutant is a severe dwarf, whereas the gid1ab and gid1bc double mutants are not (Griffiths et al., 2006; Willige et al., 2007). Conversely, GID1a and GID1b play a stronger role in fertility. Interestingly, Griffiths et al., 2006 observed that the gid1a‐1 gid1b‐1 double mutant had a significantly longer final stem length than Col‐0 WT. Thus, GID1b may also play a negative role in stem elongation. Future work should investigate whether GID1b may negatively regulate hypocotyl elongation in the dark or effects on red and far red light responses in seeds and seedlings.
Plants can evolve complex hormonal responses by having multiple hormone receptors with specialized functions. For example, it appears that only a subset of the ABA (abscisic acid) hormone receptor genes are involved in stomatal closure, making them good targets for improving drought tolerance (Okamoto et al., 2013; Park et al., 2015). The complex genetic interactions of GID1 GA receptor mutants are reminiscent of the interactions between the three salicylic acid (SA) receptors, NPR1, NPR3, and NPR4 (Fu et al., 2012; Kuai, MacLeod, & Després, 2015; Wu et al., 2012). The npr1 mutant is pathogen susceptible due to failure to induce PR (Pathogenesis Related) genes, whereas the npr3 npr4 double mutant is pathogen resistant due to increased systemic acquired resistance (SAR). The npr1 npr3 npr4 triple has an npr1‐like phenotype, indicating that NPR1 is the downstream regulator. NPR1 is a transcriptional co‐regulator that activates SAR, but represses hypersensitive response (HR). SAR allows cell survival and pathogen resistance through PR gene expression, whereas HR prevents the spread of biotrophic pathogens through programmed cell death at the initial infection site. HR needs to be limited to the infection site. Both NPR3 and NPR4 are adapter proteins for CUL3 E3 ubiquitin ligases that bind to and target NPR1 for destruction. The NPR4 receptor has a high affinity and NPR3 a low affinity for SA hormone. SA‐binding blocks the NPR4‐NPR1 interaction, but stimulates the NPR3‐NPR1 interaction. The current model describes three SA hormone conditions leading to different levels of NPR1 function. (a) In the absence of pathogen, lack of SA causes NPR4 to target NPR1 for destruction leading to lack of SAR. (b) After pathogen attack, low SA levels in cells distant from the infection site block NPR4‐directed NPR1 destruction, leading to NPR1‐stimulated SAR but not HR. (c) At the site of pathogen attack, high SA levels active NPR3‐directed NPR1 destruction, thereby lifting NPR1‐repression of HR causing cell death at the infection site. This elegant model for dose‐dependent signaling allows different responses to pathogen attack depending on SA hormone levels.
By analogy to SA signaling, different GID1 proteins may allow germination to respond to different environmental signals, possibly due to differences in GA hormone levels and/or receptor affinities (Figure 7). For example, the high GA‐affinity receptor GID1b may inhibit germination when GA levels are low (in the dark or in dormant seeds), and the lower GA‐affinity receptor GID1c may stimulate germination when GA levels are high (in the light and after‐ripened or cold‐stratified seeds) (reviewed in Yamaguchi, 2008; Nelson & Steber, 2016). GID1c is the key downstream positive regulator of light and dark germination. (Figure 5; Figure 7a,b). Based on the failure of light and dark germination in the gid1abc triple mutant, stimulation of dark germination by the gid1b and gid1a mutations requires the presence of GID1c as a positive regulator of germination. The fact that gid1b and gid1c have additive effects on dark germination of gid1bc suggests that GID1b negative regulation may be more complex, possibly functioning through an as yet unidentified target. GID1a functions independently of GID1c as a positive regulator of light and dark germination since gid1a and gid1c act additively. In light‐germinating dormant seeds, GID1b functions as an upstream negative regulator of GID1c since gid1c is epistatic to gid1b (Figure 2; Figure 7a). In the dark, GID1a and GID1b both function as negative regulators of GID1c since the gid1b and gid1ab double mutants germinate more efficiently than WT. If GID1a and GID1b did not negatively regulate germination via GID1c, then we would expect the gid1abc triple to germinate in the dark in a manner similar to gid1ab (Figure 7b). Future work will need to examine how GID1b negatively regulates GID1c. It will be interesting to learn whether the Arabidopsis GID1 genes have evolved to detect environmental conditions such as the passing of time (after‐ripening), temperature, and lighting conditions in order to ensure seedling survival after seed germination.
AUTHOR CONTRIBUTIONS
C.M.S provided the initial research design, guidance in performing and analyzing the results of experiments, and funding. W.G. obtained funding, designed and performed experiments, and performed the statistical analyses of results. C.M.S and W.G. wrote and edited the manuscript.
Supporting information
ACKNOWLEDGMENTS
The authors sincerely thank Tracy Harris for expert technical assistance and guidance. Thanks are due to Haiyan Bu for helpful discussions about seed size. The authors also thank Shantel Martinez, Nuan Wen, Scott Carle, and other members of the Steber lab for helpful advice. Thanks are due to Amber Hauvermale and Tracy Harris for critical reading of the manuscript. This research was funded by the USDA‐ARS (to CMS) and by the Chinese Scholarship Council (to WG, 201606180089).
Ge W, Steber CM. Positive and negative regulation of seed germination by the Arabidopsis GA hormone receptors, GID1a, b, and c . Plant Direct. 2018;2:1–11. 10.1002/pld3.83
References
- Ariizumi, T. , Hauvermale, A. L. , Nelson, S. K. , Hanada, A. , Yamaguchi, S. , & Steber, C. M. (2013). Lifting DELLA repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiology, 162, 2125–2139. 10.1104/pp.113.219451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, T. , Lawrence, P. K. , & Steber, C. M. (2011). The role of two F‐box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiology, 155, 765–775. 10.1104/pp.110.166272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, T. , & Steber, C. M. (2007). Seed germination of GA‐insensitive it sleepy1 mutants does not require RGL2 protein disappearance in it Arabidopsis. The Plant Cell, 19, 791–804. 10.1105/tpc.106.048009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J. D. , Bradford, K. J. , Hilhorst, H. W. M. , & Nonogaki, H . (2013). SEEDS: Physiology of development, germination and dormancy. New York, NY, Springer; 10.1007/978-1-4614-4693-4 [DOI] [Google Scholar]
- Chaisurisri, K. , Edwards, D. , & El‐Kassaby, Y. (1992). Genetic control of seed size and germination in Sitka spruce. Silvae Genet, 41, 348–355. [Google Scholar]
- Cheng, H. , Qin, L. , Lee, S. , Fu, X. , Richards, D. E. , Cao, D. , … Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development, 131, 1055–1064. 10.1242/dev.00992 [DOI] [PubMed] [Google Scholar]
- Denay, G. , Creff, A. , Moussu, S. , Wagnon, P. , Thévenin, J. , Gérentes, M.‐F. , … Ingram, G. (2014). Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix‐loop‐helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development, 141, 1222–1227. 10.1242/dev.103531 [DOI] [PubMed] [Google Scholar]
- Dill, A. , Jung, H.‐S. , & Sun, T.‐P. (2001). The DELLA motif is essential for gibberellin‐induced degradation of RGA. Proceedings of the National Academy of Sciences of the United States of America, 98, 14162–14167. 10.1073/pnas.251534098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, C. J. , & Hartwig, E. E. (1971). Effect of seed size upon rate of germination in soybeans. Agronomy Journal, 63, 429–450. 10.2134/agronj1971.00021962006300030024x [DOI] [Google Scholar]
- Ferreira, L. G. , de Alencar Dusi, D. M. , Irsigler, A. , Gomes, A. C. M. M. , Mendes, M. A. , Colombo, L. , & de Campos Carneiro, V. T. (2008). GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Reports, 37, 293–306. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R. R. , Reeves, W. , Ariizumi, T. , & Steber, C. M. (2008). Molecular aspects of seed dormancy. Annual Review of Plant Biology, 59, 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Fu, Z. Q. , Yan, S. , Saleh, A. , Wang, W. , Ruble, J. , Oka, N. , … Dong, X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature, 486, 228 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa, J. , Ito, T. , Kamiya, Y. , Yamaguchi, S. , & Takahashi, Y. (2015). Binding of GID1 to DELLAs promotes dissociation of GAF1 from DELLA in GA dependent manner. Plant Signaling & Behavior, 10, e1052923 10.1080/15592324.2015.1052923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, J. M. (2004). Bigger is not always better: Conflicting selective pressures on seed size in Quercus ilex. Evolution, 58, 71–80. 10.1111/j.0014-3820.2004.tb01574.x [DOI] [PubMed] [Google Scholar]
- Griffiths, J. , Murase, K. , Rieu, I. , Zentella, R. , Zhang, Z.‐L. , Powers, S. J. , … Thomas, S. G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in it Arabidopsis. The Plant Cell, 18, 3399–3414. 10.1105/tpc.106.047415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale, A. L. , Ariizumi, T. , & Steber, C. M. (2012). Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiology, 160, 83–92. 10.1104/pp.112.200956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale, A. L. , Ariizumi, T. , & Steber, C. M. (2014). The roles of the GA receptors GID1a, GID1b, and GID1c in sly1‐independent GA signaling. Plant Signaling & Behavior, 9, e28030 10.4161/psb.28030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale, A. L. , Tuttle, K. M. , Takebayashi, Y. , Seo, M. , & Steber, C. M. (2015). Loss of Arabidopsis thaliana seed dormancy is associated with increased accumulation of the GID1 GA hormone receptors. Plant and Cell Physiology, 56, 1773–1785. 10.1093/pcp/pcv084 [DOI] [PubMed] [Google Scholar]
- Iuchi, S. , Suzuki, H. , Kim, Y.‐C. , Iuchi, A. , Kuromori, T. , Ueguchi‐Tanaka, M. , … Nakajima, M. (2007). Multiple loss‐of‐function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. The Plant Journal, 50, 958–966. 10.1111/j.1365-313X.2007.03098.x [DOI] [PubMed] [Google Scholar]
- King, K. E. , Moritz, T. , & Harberd, N. P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics, 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai, X. , MacLeod, B. J. , & Després, C. (2015). Integrating data on the Arabidopsis NPR1/NPR3/NPR4 salicylic acid receptors; a differentiating argument. Frontiers in Plant Science, 6, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Cheng, H. , King, K. E. , Wang, W. , He, Y. , Hussain, A. , … Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA‐like gene whose expression is up‐regulated following imbibition. Genes & Development, 16, 646–658. 10.1101/gad.969002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman, M. R. , Wright, I. J. , Moles, A. T. , & Westoby, M . (2000). Chapter 2: The evolutionary ecology of seed size In Fenner M. (Ed), Seeds: The ecology of regeneration in plant communities (pp. 31–57). New York, NY: CABI; 10.1079/9780851994321.0000 [DOI] [Google Scholar]
- McKinney, E. C. , Ali, N. , Traut, A. , Feldmann, K. A. , Belostotsky, D. A. , McDowell, J. M. , & Meaghr, R. B. (1998). The Arabidopsis SLEEPY1 gene encodes a putative F‐box subunit of an SCF E3 ubiquitin ligase. The Plant Cell, 15, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K. M. , Thomas, S. G. , Soule, J. D. , Strader, L. C. , Zale, J. M. , Sun, T.‐P. , & Steber, C. M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F‐box subunit of an SCF E3 ubiquitin ligase. The Plant Cell, 15, 1120–1130. 10.1105/tpc.010827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M. , Shimada, A. , Takashi, Y. , Kim, Y.‐C. , Park, S.‐H. , Ueguchi‐Tanaka, M. , … Yamaguchi, I. (2006). Identification and characterization of Arabidopsis gibberellin receptors. The Plant Journal, 46, 880–889. 10.1111/j.1365-313X.2006.02748.x [DOI] [PubMed] [Google Scholar]
- Nelson, S. K. , Ariizumi, T. , & Steber, C. M. (2017). Biology in the dry seed: Transcriptome changes associated with dry seed dormancy and dormancy loss in the Arabidopsis GA‐insensitive sleepy1‐2 mutant. Frontiers in Plant Science, 8, 2158 10.3389/fpls.2017.02158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S. K. , & Steber, C. M. (2016). Gibberellin hormone signal perception: Down‐regulating DELLA repressors of plant growth and development In Hedden P. & Thomas S.G. (Eds.), “The Gibberellins” (pp. 153–188). Hoboken, NJ: Wiley‐Blackwell, Annual Plant Reviews 49 [Google Scholar]
- Okamoto, M. , Peterson, F. C. , Defries, A. , Park, S.‐Y. , Endo, A. , Nambara, E. , … Cutler, S. R. (2013). Activation of dimeric ABA receptors elicits guard cell closure, ABA‐regulated gene expression, and drought tolerance. Proceedings of the National Academy of Sciences of the United States of America, 110, 12132–12137. 10.1073/pnas.1305919110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.‐Y. , Peterson, F. C. , Mosquna, A. , Yao, J. , Volkman, B. F. , & Cutler, S. R. (2015). Agrochemical control of plant water use using engineered abscisic acid receptors. Nature, 520, 545 10.1038/nature14123 [DOI] [PubMed] [Google Scholar]
- Thomas, S. G. , Blázquez, M. A. , & Alabadi, D. (2016). DELLA proteins: Master regulators of gibberellin responsive growth and development In Hedden P. & Thomas S.G. (Eds.), “The Gibberellins” (pp. 189–228). Wiley‐Blackwell; Annual Plant Reviews, 49. [Google Scholar]
- Tyler, L. , Thomas, S. G. , Hu, J. , Dill, A. , Alonso, J. M. , Ecker, J. R. , & Sun, T.‐P. (2004). DELLA proteins and gibberellin‐regulated seed germination and floral development in Arabidopsis. Plant Physiology, 135, 1008–1019. 10.1104/pp.104.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi‐Tanaka, M. , Ashikari, M. , Nakajima, M. , Itoh, H. , Katoh, E. , Kobayashi, M. , … Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature, 437, 693–698. 10.1038/nature04028 [DOI] [PubMed] [Google Scholar]
- Urbanova, T. , & Leubner‐Metzger, G . (2016). Chapter 9: Gibberellins and Seed germination In Hedden P. & Thomas S.G. (Eds.), The Gibberellins. Annual Plant Reviews Volume 49. Oxford, UK: Wiley‐Blackwell Publishing. [Google Scholar]
- Voegele, A. , Linkies, A. , Mueller, K. , & Leubner‐Metzger, G. (2011). Members of the gibberellin receptor gene family it GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during it Lepidium sativum and it Arabidopsis thaliana seed germination. Journal of Experimental Botany, 62, 5131–5147. 10.1093/jxb/err214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , & Deng, X. W. (2011). Plant ubiquitin‐proteasome pathway and its role in gibberellin signaling. Cell Research, 21, 1286 10.1038/cr.2011.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild, M. , Davière, J.‐M. , Cheminant, S. , Regnault, T. , Baumberger, N. , Heintz, D. , … Achard, P. (2012). The Arabidopsis DELLA RGA‐LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. The Plant Cell, 24, 3307–3319. 10.1105/tpc.112.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B. C. , Ghosh, S. , Nill, C. , Zourelidou, M. , Dohmann, E. M. N. , Maier, A. , & Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. The Plant Cell, 19, 1209–1220. 10.1105/tpc.107.051441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , & Du, G. (2007). Germination is related to seed mass in grasses (Poaceae) of the eastern Qinghai‐Tibetan Plateau, China. Nordic Journal of Botany, 25, 361–365. 10.1111/j.0107-055X.2007.00179.x [DOI] [Google Scholar]
- Wu, Y. , Zhang, D. , Chu, J. Y. , Boyle, P. , Wang, Y. , Brindle, I. D. , … Després, C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports, 1, 639–647. 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annual Review of Plant Biology, 59, 225–251. 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y. , Hirai, T. , Yamamoto, E. , Kawamura, M. , Sato, T. , Kitano, H. , … Ueguchi‐Tanaka, M. (2010). A rice it gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. The Plant Cell, 22, 3589–3602. 10.1105/tpc.110.074542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Ito, T. , Zhao, Y. , Peng, J. , Kumar, P. , & Meyerowitz, E. M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proceedings of the National Academy of Sciences of the United States of America, 101, 7827–7832. 10.1073/pnas.0402377101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


