Abstract
The proficiency of cancer cells to repair DNA double-strand breaks (DSBs) by homologous recombination (HR) is a key determinant in predicting response to targeted therapies such as PARP inhibitors. The RAD51 paralogs work as multimeric complexes and act downstream of BRCA1 to facilitate HR. Numerous epidemiological studies have linked RAD51 paralog mutations with hereditary cancer predisposition. Despite their substantial links to cancer, RAD51 paralog HR function has remained elusive. Here we identify isoform 1 as the functional isoform of RAD51D, whereas isoform 4 which has a large N-terminal deletion (including the Walker A motif), and isoform 6 which includes an alternate exon in the N-terminus, are non-functional. To determine the importance of this N-terminal region, we investigated the impact of cancer-associated mutations and SNPs in this variable RAD51D N-terminal region using yeast-2-hybrid and yeast-3-hybrid assays to screen for altered protein-protein interactions. We identified two cancer-associated mutations close to or within the Walker A motif (G96C and G107V, respectively) that independently disrupt RAD51D interaction with XRCC2. We validated our yeast interaction data in human U20S cells by co-immunoprecipitation and determined the impact of these mutations on HR-proficiency using a sister chromatid recombination reporter assay in a RAD51D knock-out cell line. Our investigation reveals that the interaction of RAD51D with XRCC2 is required for DSB repair. By characterizing the impact of cancer-associated mutations on RAD51D interactions, we aim to develop predictive models for therapeutic sensitivity and resistance in patients who harbor similar mutations in RAD51D.
Keywords: Homologous recombination, RAD51 paralogs, RAD51D, XRCC2, Double-strand break repair, Walker A motif
1. Introduction
HR is a high-fidelity DSB repair mechanism that utilizes a homologous template for repair [1]. Cells commit to HR by resecting the broken DNA ends to yield a single-stranded DNA (ssDNA) overhang, which becomes coated with Replication Protein A (RPA). RPA is then exchanged for RAD51 recombinase to form a RAD51 nucleoprotein filament. RAD51 filaments perform the homology search and strand invasion steps of HR [2]. RAD51 filament formation is aided by the RAD51 paralogs, which structurally resemble RAD51 and if depleted, impair HR [3].
There are six mammalian RAD51 paralogs including RAD51B, RAD51C, RAD51D, XRCC2, XRCC3 and SWSAP1 [4, 5]. These proteins form distinct sub-complexes including BCDX2 (RAD51B, RAD51C, RAD51D and XRCC2), CX3 (RAD51C and XRCC3) and the Shu complex (SWSAP1 and SWS1) [4, 6, 7]. The RAD51 paralogs proteins have roles in DNA integrity, including replication stress tolerance by promoting DNA replication fork stability and restart, DNA DSB repair by promoting RAD51 filament formation, and in the case of RAD51D, telomere length maintenance [8–10]. The diverse requirements for these proteins are also exemplified in yeast, where the RAD51 paralogs promote error-free damage tolerance of base-excision repair intermediates following DNA alkylation as well as efficient DNA DSB repair [11]. Despite substantial evidence linking human RAD51 paralog mutations with breast and ovarian cancer, their function during DNA repair and the impact of specific cancer-associated mutations remains largely unknown [12–17].
Due to the early embryonic lethality observed in RAD51D knock-outs in rodents, researchers used Rad51d-deficient Chinese hamster ovary cell lines to determine the importance of RAD51D and found that knock-outs exhibited an increased loss of genomic DNA resulting from use of the alternative single-strand annealing pathway [18, 19]. Like RAD51, the RAD51D and XRCC2 contain canonical Walker A and Walker B motifs, characteristic of proteins with ATPase activity [20, 21]. Despite this, whether or not ATPase activity of RAD51D is required for DNA repair remains controversial. For example, two reports suggest different requirements for the RAD51D Walker A motif for its XRCC2 interaction and protein function [20, 22].
Studies characterizing RAD51 paralog structure and function have been plagued by poor protein solubility, low protein stability, and profound sickness/lethality observed in knock-out model systems [19, 23–25]. In addition, it remains unknown which human RAD51D isoform is responsible for mediating its HR functions. Here we determine that RAD51D isoform 1 is the RAD51D isoform able to restore HR in a RAD51D knock-out background. We then employed the powerful yeast-2-hybrid (Y2H) and yeast-3-hybrid (Y3H) systems to study the effect of cancer-associated mutations and population SNPs in RAD51D on its interaction with XRCC2. We identified two glycine residues in close proximity to each other upstream and part of the Walker A motif (G96 and G107, respectively) that are required for RAD51D interaction with XRCC2. We validated our Y2H findings by co-immunoprecipitation in human U2OS cells. Importantly both the G96C and G107V RAD51D mutants alone or in combination exhibit reduced HR, using a GFP-based sister chromatid recombination (SCR) reporter assay [26]. Finally, we explored the possibility that the protein-protein interactions of RAD51D, and ultimately its function, may be controlled by post-translational modifications in this region.
2. Materials and methods
2.1. Tissue culture, cell lines and reagents
Human U20S and U20S SCR (Sister chromatid recombination) #18 Wild-Type (WT) [26] and RAD51D CRISPR Knock-out (Clone D4) cell lines (both gifted from Mauro Modesti; Garcin et al., in preparation) were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin and streptomycin. Cells were transfected using TranslT®-LT1 transfection reagent (Mirus Bio) diluted in OptiMEM® serum free media and following the manufacturer’s instructions. RAD51D cDNAs were expressed from mammalian expression plasmids pCDNA3 FLAG-RAD51D isoform 1 (NM_002878.3) (gifted from Paul Russell [27]), pCDNA3.1 3xFLAG-RAD51 D isoform 4 (NM_133629.2) and isoform 6 (NM_001142571.1) (supplied by Novoprolabs). pCBAScel was a gift from Maria Jasin (Addgene plasmid # 26477) [28], Mutations were introduced into the cDNA of RAD51D by site-directed mutagenesis using Phusion polymerase master mix (M0531S, NEB) and Dpnl (R0176S, NEB). All mutations were verified by DNA sequencing (Genewiz).
2.2. Sister chromatid recombination reporter assay
U20S SCR #18 WT and RAD51D Knock-out (D4) cells were seeded into a single well of a 6-well plate at 1×105 and 2×105 cells per well respectively (due to slower growth of cells lacking RAD51D). Cells were incubated for 24 hours before co-transfection with an l-Scel expressing plasmid and either an empty vector control or a vector expressing FLAG-RAD51D using Mirus LT1 transfection reagent at a ratio of 1:2 of plasmid DNA to transfection reagent. For co-transfections, a total of 5pg of plasmid DNA was transfected per well in a 1:1 ratio of l-Scel to Empty vector or FLAG-RAD51D). Transfection efficiency was assessed by transfection of a separate sample of U20S cells with eGFP.
Following transfection, cells were incubated for a further 48 hours before trypsinization and analysis by fluorescence activated cell sorting. For each assay, 50,000 events per experiment were analyzed in three experiments. To account for variable transfection efficiency between experiments, results were normalized to one additional sample per experiment transfected solely with pEGFP.
2.3. Yeast-2-hybrid and yeast-3-hybrid assays
For yeast-2-hybrid assays, a pGBD-RAD51D vector was used to express RAD51D C-terminally fused to the DNA-binding domain of GAL4 and a pGAD-XRCC2 vector was used to express XRCC2 C-terminally fused to the GAL4-activation domain. PJ69-4A cells were co-transformed with the above plasmids and plated on synthetic complete medium lacking leucine and tryptophan (SC-Leu¯Trp¯). Single-colonies were selected and grown overnight. The following morning cultures were diluted to 0.2 OD600 and cultured for a further 3 hours (until ~0.5 OD600) equal numbers of cells were spotted in 5μl of culture medium onto synthetic complete medium lacking leucine, tryptophan and histidine (SC-Leu¯Trp¯His¯; least stringent), with 3-amino-1,2,4-triazole (SC-Leu¯Trp¯His¯+3-AT; more stringent) or synthetic complete medium lacking leucine, tryptophan, histidine and adenine (SC-Leu¯Trp¯His¯Ade¯; most stringent). Plates were incubated for 48 hours at 30 °C before taking images. All images were adjusted for brightness and contrast identically using Adobe Photoshop.
For yeast 3-hybrid experiments, pGBD-RAD51D, pGAD-RAD51C and pRS416-RAD51B were co-transformed into PJ69-4A and transformed onto synthetic complete medium lacking leucine, tryptophan and uracil (SC-Leu¯Trp¯Ura¯). As before, single-colonies were selected and grown overnight. The following morning cultures were diluted to 0.2 OD600 and cultured for a further 3 hours (until ~0.5 OD600) equal numbers of cells were spotted in 5μl of culture medium on either SC-Leu¯Trp¯Ura¯His¯medium; least stringent, SC-Leu¯Trp¯Ura¯His¯+3-AT medium; more stringent or SC-Leu¯Trp¯Ura¯His¯Ade¯medium; most stringent. Medium with SC-Leu¯Trp¯Ura¯was used as a loading control. Plates were incubated for 48 hours at 30 °C before taking images. All images were adjusted for brightness and contrast identically using Adobe Photoshop.
2.4. Co-immunoprecipitation
U20S Cells were incubated for 24 hours after transfection to express F1_AG-RAD51 D and Myc-XRCC2. Cells were harvested by scraping into cold phosphate-buffered saline (PBS) before lysis in lysis buffer supplemented with protease inhibitors (A32965, Pierce™), phosphatase inhibitors (A32957, Pierce™) and 2% (v/v) Benzonase (70746, Millipore Sigma). Lysis buffer: 50mM Tris base, 100mM NaCI, 2mM MgCh, 10% glycerol, 0.1% NP-40, 1mM DTT. Cell lysates were incubated for 30 minutes on ice before clearing by centrifugation and incubation overnight with c-Myc (9E10) agarose (sc-40 AC, Santa Cruz). The next day, the agarose beads were washed four times with fresh lysis buffer before addition of SDS-sample buffer and boiling for 5 minutes. Input and elution samples were run on SDS-PAGE and analyzed by Western blotting (as described in 2.5). All images were adjusted for brightness and contrast identically using Adobe Photoshop.
2.5. Western blot analyses
Samples harvested for Western blot analysis were run on 10% SDS-PAGE gels. Proteins were transferred to PVDF membrane (lmmobilon®-FL, Millipore) before blocking for 1 hour with Odyssey® blocking buffer (927-50000, Licor®). Antibodies were diluted in a 1:1 ratio with (Tris phosphate buffer) TBS and Odyssey® blocking buffer supplemented with 0.2% Tween-20. Antibodies against Fl_AG-M2 (F3165, Sigma), c-Myc (sc-789, Santa Cruz), RAD51D (sc-398819, Santa Cruz) and α-Tubulin (2144, Cell Signalling) were all used at a 1:1,000 dilution. Antibodies against XRCC2 (TA327180, OriGene) was used at a 1:2,000 dilution. Secondary antibodies, IRDye®680RD antirabbit (926-68071, Licor®) and IRDye®800CW (926-32210, Licor®), were used at 1:20,000. Membranes were imaged using a Licor Odyssey® CLx imaging system.
3. Results and discussion
3.1. RAD51D splicing isoform 1, but not isoforms 4 or 6, are required for repair of DSBs by HR
Upon the discovery of the murine and human RAD51D, it was noted that several splicing isoforms exist [29], Later investigation identified 7 RAD51D splice variants in both mice and humans. In humans, four of the RAD51D isoforms are only found in brain tissue (RAD51DΔ3,5 RAD51DΔ5, RAD51DΔ4,5 and RAD51D+int3Δ4,5) whereas the other isoforms are differentially expressed in several tissues including brain, kidney, mammary and lung tissues as well as in the testis, cervix, prostate and spleen (RAD51DΔ3, RAD51DΔ3,4,5 and RAD51D+int3 also referred to as transcript variants 1, 4 and 6, respectively) [30], Investigation into the protein-protein interactions of the murine isoforms of RAD51D cells revealed that in mice, RAD51D isoform Δ7b is proficient for interactions with both binding partners, RAD51C and XRCC2, with a second murine isoform (RAD51D+int3; synonymous with human RAD51D splicing isoform 6) is only proficient for XRCC2 binding [30]. Interestingly, both isoforms fail to rescue sensitivity of Rad51d-deficient cells to interstrand crosslinking agents [30]. We cannot exclude the possibility that these splicing isoforms may have altered protein stability, cellular localization or DNA-binding capability. It is also possible that expression of these alternate transcripts may play a regulatory role in DNA repair by competing with full-length RAD51D for interactions with its binding partners, RAD51C and XRCC2.
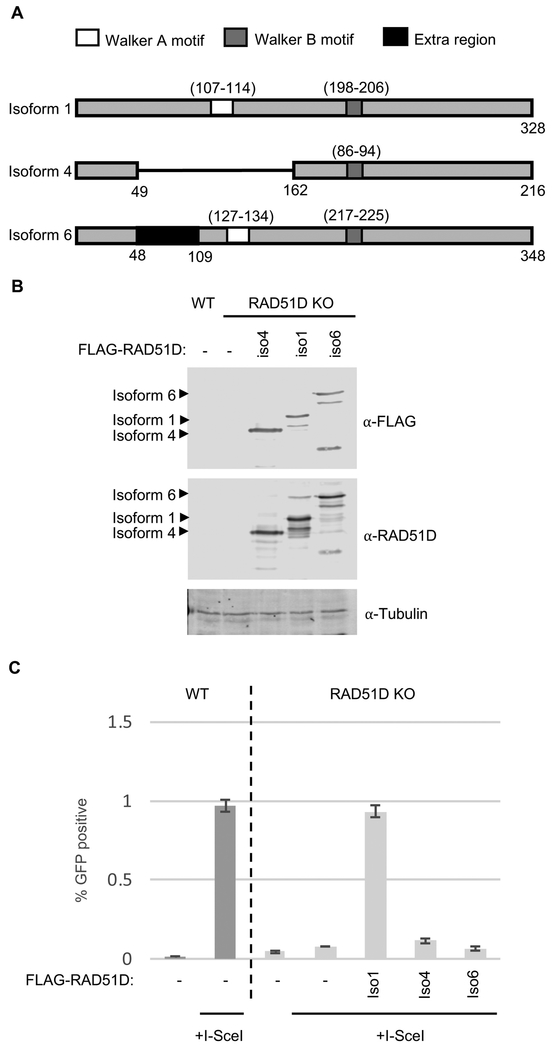
To investigate the function of human RAD51D in response to DSBs, we first aimed to determine which human RAD51D isoform is functional. We identified three coding isoforms of human RAD51D, transcript variants 1, 4 and 6 from the nucleotide database provided by NCBI [31]. Figure 1A shows a schematic of the three isoforms tested. Transcript isoform 1 contains both Walker A and B motifs and is proposed to be the functional full-length protein at 328 amino acids in length. Transcript isoform 4 is shorter at 216 amino acids in length, lacking exons 3, 4 and 5 of exons 1-10 resulting in the loss of a portion of the N-terminal region including the Walker A motif from the final protein product. Transcript isoform 6 includes an alternate exon 3 resulting in an extra region adjacent to the Walker A motif. This isoform is longer than isoform 1 at 348 amino acids in length. Each of these isoforms can be N-terminally FLAG-tagged and are readily expressed in U2OS RAD51D knock-out cell lines (Figure 1B; Supplemental Figure 1A). We note that some degradation products are present in the cells expressing exogenous RAD51D. It is well characterized that RAD51 paralogs do not express efficiently without reciprocal binding partners to aid their protein stability and solubility [6]. Despite this, we are able to observe expression of each of these isoforms above endogenous RAD51D protein levels.
Figure 1. RAD51D splicing isoform 1, but not isoforms 4 or 6, rescues HR in RAD51D knock-out U20S cell lines.
A) Schematic diagram of the three coding isoforms of RAD51D (isoforms 1,4 and 6). Amino acid residue numbers of the Walker A (white box) and B (dark grey box) motifs are shown in parenthesis. The extra coding region in isoform 6 is shown in black.
B) U20S wild-type or RAD51D knock-out cells were seeded on 35mm plates 24-hours prior to transfection with FLAG-RAD51D cDNA constructs. Following transfection, cells were incubated for an additional 24-hours before lysis and Western blot analysis for FLAG expression using α-Flag antibodies. α-Tubulin was used as a loading control.
C) Cells were seeded 24-hours prior to transfection as in B. Cells were co-transfected with I-Scel and FLAG-RAD51D cDNAs or an empty vector control. Cells were incubated for an additional 48-hours and then analyzed by flow cytometry for green-fluorescence. The bar chart shows the average percentage of GFP+ cells over three experiments. Error bars show one standard deviation from the mean.
To examine whether each isoform was DSB repair proficient, we used a recombination reporter assay integrated into wild-type and RAD51D knock-out U2OS cells (U2OS SCR clone 18; [26] Garcin et al., in preparation). This assay uses a GFP coding gene disrupted with an I-Scel restriction site as well as an incomplete downstream GFP copy that can be used for templated repair. If the DSB is repaired by HR, a functional GFP gene is restored and can be analyzed by flow cytometry. Analysis of GFP positive cells following co-transfection of I-Scel with either RAD51D isoform 1, 4 or 6 in a RAD51D knock-out U2OS cell reveals that only RAD51D isoform 1 restores HR following DSB induction (Figure 1C).
Both RAD51D splice variants, isoforms 4 and 6, only differ in their N-terminus including and/or adjacent to the Walker A motif. We hypothesized that mutations within this region may be critical for RAD51D function. Previous studies defining the binding regions of RAD51D with XRCC2 and RAD51C, demonstrated that the murine RAD51D fragment (amino acids 4-77) is proficient for its XRCC2 interaction but not with RAD51C. Gruver et al (2009) similarly found that amino acids 54-77 are indispensable for murine RAD51D interaction with XRCC2 [30]. In contrast, a RAD51D C-terminal fragment (amino acids 77-328) is proficient for its RAD51C interaction, but not with XRCC2 [32]. Taken together, it is possible that RAD51D isoforms 4 and 6 do not interact with XRCC2 due to the disruption or absence of this region. Unfortunately, we are unable to test this hypothesis because XRCC2 failed to efficiently express in cells co-transfected with RAD51D isoforms 4 and 6, potentially indicating a RAD51 paralog complex stability impairment. Therefore, it is possible that specific RAD51D point mutations in this region could be used to disrupt the interaction of RAD51D with its binding partners, XRCC2 and RAD51C.
3.2. Identification of cancer-associated RAD51D mutations, G96C and G107V, impair its interaction with XRCC2 and RAD51C in a yeast-2-hybrid and yeast-3-hybrid system, respectively
Having established that RAD51D transcript isoform 1 is the functional isoform required for DSB repair, we next examined RAD51D’s protein interactions with the other RAD51 paralogs using a Y2H approach. Similar to what has been reported [33], RAD51 paralog protein-protein interactions can be readily observed by Y2H (Figure 2A and 2B). Previous work demonstrated that RAD51D, as a member of the BCDX2 complex, directly interacts with both XRCC2 and RAD51C [6, 34], Each of these interactions can be readily assayed in the Y2H (XRCC2) or Y3H (RAD51C) system (Figure 2). Note that RAD51D interaction with RAD51C is examined using a Y3H approach because RAD51B helps stabilize RAD51C expression (Figure 2E; [22]). We find that RAD51D exhibits the strongest Y2H interaction with XRCC2 and does not exhibit a Y2H interaction with XRCC3, RAD51B, SWSAP1, or SWS1 (Figure 2A). Therefore, Y2H and Y3H assays can be used to quickly screen RAD51D mutations to determine whether any of the observed protein-protein interactions are affected.
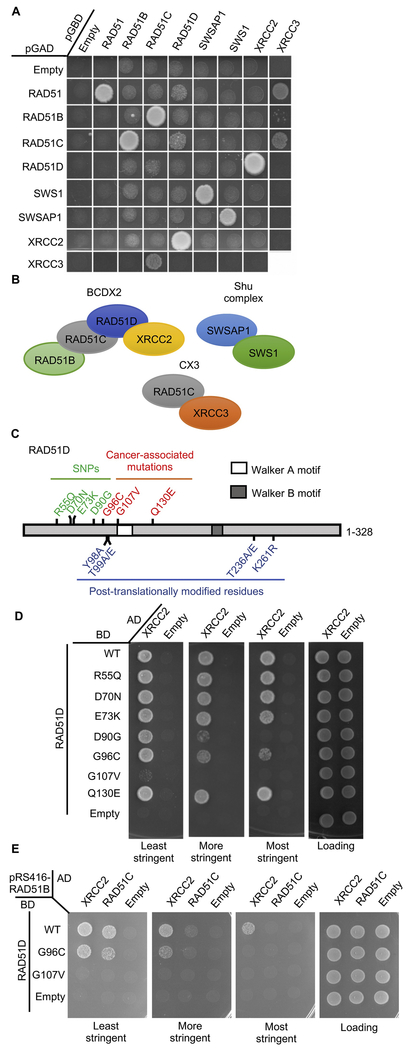
Figure 2. Identification of cancer-associated mutations, G96V and G107V, in RAD51D that impair its interaction with XRCC2 and RAD51C.
A) Yeast 2-hybrid analysis showing the interactions of each of the RAD51 paralogs (and SWS1, a SWIM-domain containing protein) with each other. The PJ694a yeast strain was transformed with a plasmid where RAD51, the RAD51 paralogs (RAD51B, RAD51C RAD51D, SWSAP1, XRCC2, and XRCC3) and SWS1 were cloned into a plasmid that expresses the GAL4 DNA activating (pGAD) and GAL4 DNA binding (pGBD) domains.A yeast-2-hybrid interaction was assayed by platting the yeast on SC-Leu¯Trp¯His¯ and compared to the empty pGAD or pGBD plasmids, which were used as negative controls. Note that XRCC3 analysis had to be performed on a separate plate because of the plate size and that, consistent with the yeast Rad51 paralogs, human RAD51 Y2H interactions with the RAD51 paralogs are only observed when RAD51 is expressed in the pGAD vector [57].
B) Diagram representing the protein-protein interaction between the RAD51 paralog subcomplexes shown in the part A. BCDX2 is composed of RAD51B (light green), RAD51C (grey), RAD51D (blue), and XRCC2 (yellow). The CX3 complex is composed of RAD51C (grey) and XRCC3 (orange) and the Shu complex composed of SWSAP1 (light blue) and SWS1 (green).
C) Schematic diagram showing RAD51D Walker motifs (white and dark grey boxes) as well as SNPs (green), cancer-associated mutations (red) and potentially post-translationally modified residues (blue) analyzed.
D) Yeast-2-hybrid analysis of SNPs and cancer-associated mutations in RAD51D on its interaction with XRCC2. The PJ694a yeast strain was transformed with a plasmid where RAD51D or the indicated RAD51D mutation was fused to the GAL4-DNA binding domain [BD; pGBD-RAD51D (WT)] and a plasmid where XRCC2 was fused to the GAL4-DNA activating domain (AD; pGAD-XRCC2). A yeast-2-hybrid interaction was assayed by platting the yeast on SC-Leu¯Trp¯His¯ (least stringent), SC-Leu¯Trp¯His¯+3AT (more stringent), or SC-Leu¯Trp¯His¯Ade¯ (most stringent) and compared to the loading control (SC-Leu¯Trp¯). Empty AD (pGAD; Empty) plasmid was used as a negative control.
E) Yeast-3-hybrid analysis of RAD51D-G96C and/or -G107V mutations with XRCC2 and RAD51C. The PJ694a yeast strain was transformed with three plasmids; 1) a plasmid where RAD51D (or the indicated mutant) was fused to the GAL4-DNA binding domain [BD; pGBD-RAD51D (WT)], 2) a plasmid where either XRCC2 or RAD51C was fused to the GAL4-DNA activating domain (AD; pGAD-XRCC2, pGAD-RAD51C), and 3) a plasmid that constitutively expressed RAD51B (pRS416-RAD51B). Note that a plasmid containing RAD51B was used to help stabilize RAD51C expression. A yeast-3-hybrid interaction between RAD51D with XRCC2 or RAD51C was assayed by platting the yeast on SC-Leu¯Trp¯Ura¯His¯ (least stringent), SC-Leu¯Trp¯Ura¯His¯+3AT (more stringent), or SC-Leu¯Trp¯Ura¯His¯Ade¯ (most stringent) and compared to the loading control SC-Leu¯Trp¯Ura¯ (Loading). Empty pGBD (BD Empty) and pGAD (AD Empty) plasmids were used as negative controls.
We next wanted to screen cancer-associated mutations and germline SNPs in the N-terminal half of RAD51D for altered protein-protein interactions. To do this, we used the publicly available COSMIC and EXOME databases [Table 1; schematic of mutations shown in red (cancer-associated) and green (SNPs) in Figure 2C]. Due to the high number of cancer-associated mutations, several mutations in the N-terminus of RAD51D were selected using the PolyPhen-2 server (Table 2). PolyPhen-2 predicts the likelihood of an amino acid substitution mutation having an impact on the structure and/or function of a protein based on the gene structural and evolutionary conservation [35], Based on this information, the PolyPhen-2 generates a score between (0 and 1) with a score of 1 being an amino acid substitution most likely to be damaging to the proteins function. We introduced substitution mutations into the cDNA of RAD51D that were predicted to be ‘possibly damaging’ or ‘probably damaging’ prior to screening for altered Y2H interactions. The use of ‘possibly damaging’ and ‘probably damaging’ classifications is used to specify the confidence in the prediction based on the false positive rate thresholds set by the server.
Table 1.
- RAD51D mutations identified in EXOME and COSMIC databases. Table shows which mutations are germline SNPs (green) and which are somatic cancer-associated mutations (red). Table details the tissue in which the mutation was originally identified.
| Mutation | Tissue origin | Database | Somatic vs Germ line | References |
|---|---|---|---|---|
| R55Q | Not Applicable | EXOME | Germline | [36] |
| D70N | Not Applicable | EXOME | Germline | [36] |
| E73K | Not Applicable | EXOME | Germline | [36] |
| D90G | Not Applicable | EXOME | Germline | [36] |
| G96C | Ovary | Ν/Α | Germline | [37] |
| G107V | Lung (Small Cell Carcinoma) | COSMIC | Somatic | [38, 39] |
| Q130E | Lung (Squamous Cell Carcinoma) | COSMIC | Somatic | [40] |
Table 2.
- RAD51D PolyPhen-2 analysis. RAD51D isoform 1 was entered in FASTA format along with the amino acid substitution and position information. Mutation scores were generated along with a prediction of whether the mutation would be benign, possibly or probably damaging. Mutations analyzed were selected on the basis of the mutation being ‘possibly damaging’ or ‘probably damaging’.
| Mutation | Score | Prediction |
|---|---|---|
| R55C | 0.936 | Possibly Damaging |
| D70N | 0.933 | Possibly Damaging |
| E73K | 0.9 | Possibly Damaging |
| D90G | 0.735 | Possibly Damaging |
| G96C | 0.999 | Probably Damaging |
| G107V | 1 | Probably Damaging |
| C130E | 0.999 | Probably Damaging |
Of the seven cancer-associated mutations and SNPs in RAD51D screened, only the RAD51D-G107V mutation exhibited a strongly impaired Y2H interaction with XRCC2 (Figure 2D). RAD51D-D90G and RAD51D-G96C exhibited modest XRCC2 Y2H impairment upon the higher stringency plates (Figure 2D; more and most stringent plates). To further investigate this, we introduced the RAD51D-G107V and the nearby G96C mutation into the Y3H system to test their interaction with RAD51C. We find that RAD51D-G107V exhibits impaired interaction with both RAD51C and XRCC2. Note that the RAD51D-G96C mutant exhibited a modest Y3H impairment upon the higher stringency plates (Figure 2E). Together these results suggest that G96 and G107 may be important residues in RAD51D for its protein interactions.
3.3. Cancer-associated RAD51D mutations, G96C, G107V and the double-mutant G96C/G107V, exhibit impaired XRCC2 interaction and HR-proficiency in human cells
Following identification of RAD51D cancer-associated mutations that disrupt its XRCC2 interaction in yeast, we next aimed to validate these findings in mammalian cells. To do this, we first determined that the mutant RAD51D proteins (RAD51D-G96C, RAD51D-G107V, and RAD51D-G96C/G107V) are stably expressed in human U20S cells following transfection (Figure 3A). Next, we performed co-immunoprecipitation experiments by co-transfecting either wild-type FLAG-RAD51D or mutant FLAG-RAD51D with Myc-XRCC2. In contrast to wild-type RAD51D, both RAD51D-G96C and RAD51D-G107V mutants exhibit an impaired XRCC2 interaction (Figure 3B). Note that the Y2H system enables examination of direct protein interactions, whereas in human cells, these interactions may be stabilized by other binding proteins. This likely accounts for the partial deficiency observed in the RAD51D-XRCC2 interaction by co-immunoprecipitation. To fully investigate the potential impact of these mutants in human cells, we also generated a double RAD51D mutant (RAD51D-G96C/G107V). The RAD51D double-mutant further decreased the ability of RAD51D to co-immunoprecipitate with XRCC2, indicating that both of these residues independently contribute to the RAD51D-XRCC2 protein interaction.
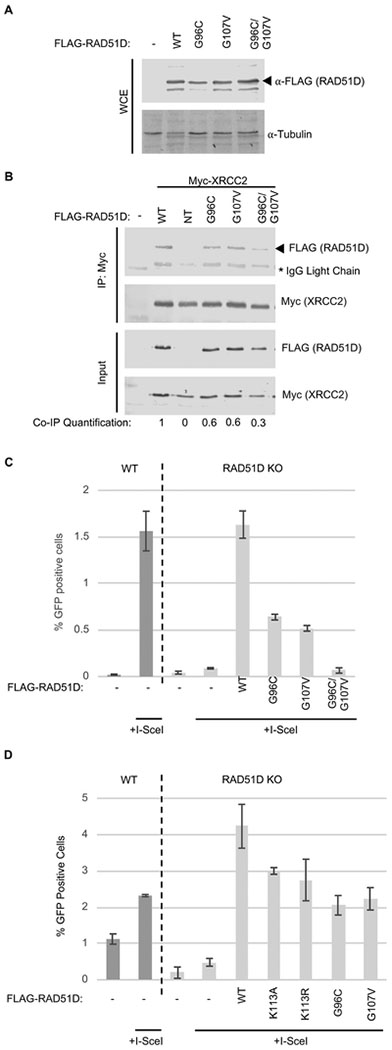
Figure 3. RAD51D-G96C, -G107V and the double -G96C/G107V mutant exhibit impaired XRCC2 interaction and HR in human U20S cells.
A) U20S wild-type or RAD51D knock-out cells were seeded on 35mm plates 24-hours prior to transfection with FLAG-RAD51D mutant cDNA constructs. Following transfection, cells were incubated for a further 24-hours before lysis and Western blot analysis for FLAG expression using aFLAG antibodies. α-Tubulin was used as a loading control.
B) Co-immunoprecipitation of Myc-XRCC2 with FLAG-RAD51D-G96C, G107V and the double-mutant from human U20S cells. U20S cells were seeded on 100mm plates 24-hours prior to transfection with FLAG-RAD51D wild-type or mutant cDNA constructs (NT= not transfected). Following transfection, cells were incubated for an additional 24-hours before lysis. Lysates were incubated overnight at 4°C with α-Myc agarose beads. The next day, the agarose beads were washed and the co-immunoprecipitates were eluted using SDS-sample buffer. Samples were analyzed by Western blotting for a- FLAG (co-immunoprecipitation) and α-Myc (immunoprecipitation). Western blot band densitometry was calculated using Image Studio software, co-immunoprecipitation FLAG-band intensity (Co-IP Quantification) was normalized to immunoprecipitation (IP: Myc) and to input FLAG expression to account for transfection efficiency and any variability in the immunoprecipitation. *Signifies the bands produced by cross-reactivity of the secondary antibody with IgG light-chain of the Myc antibody used for immunoprecipitation.
C) U20S wild-type or RAD51D knock-out cells were seeded on 35mm plates 24-hours prior to transfection with the indicated FLAG-RAD51D cDNA mutant constructs. Cells were co-transfected with either l-Scel and FLAG-RAD51D cDNAs or l-Scel and an empty vector control. Cells were incubated for an additional 48-hours and analyzed by flow cytometry for green-fluorescence. The bar chart shows the average percentage of GFP+ cells over three experiments. Error bars show one standard deviation from the mean.
D) U20S wild-type or RAD51D knock-out cells were seeded and transfected as in 3C (including K113A and K113R; Walker A motif mutants). Cells were incubated for an additional 48-hours and analyzed by flow cytometry for green-fluorescence. The bar chart shows the average percentage of GFP+ cells over three experiments. Error bars show one standard deviation from the mean
Since we identified important residues in RAD51D for its XRCC2 interaction, we next examined whether impairing RAD51D’s interaction with XRCC2 would result in impaired HR. To test this, we complemented a RAD51D knockout cell line containing a recombinational reporter assay with either RAD51D wild-type or the G96C, G107V or the double-mutant (Figure 3C). We find that both single-mutations failed to fully rescue HR in the RAD51D knock-out cell lines, producing a ~65% decrease in HR events (Figure 3C). This HR defect was further compounded when cells were transfected with the RAD51D double-G96C/G107V-mutant (Figure 3C). These results suggest that the interaction between RAD51D with its binding partner XRCC2 is important for RAD51D’s HR function.
Following the identification of mutants adjacent to the RAD51D Walker A motif, we sought to clarify whether or not the Walker A is dispensable for DSB repair by HR. Despite varied reports regarding a functional requirement of the Walker A motif in tolerating MMC-induced DNA damage [20, 22], we find that the expression of RAD51D containing point mutations, K113A and K113R, fails to fully restore HR proficiency in RAD51D knock-out U20S cells (Figure 3D). This suggests that although K113A/R mutants do not disrupt the RAD51D-XRCC2 interaction [20, 22], there is a partial requirement for the RAD51D Walker A K113 residue, at least in the context of DSB repair by HR.
3.4. Mutation of N-terminal residues that are potentially post-translationally modified do not affect RAD51D protein-protein interactions in yeast or HR-proficiency in human cells
RAD51 paralog function is regulated through protein-protein interactions and also through post-translational modifications. For example, XRCC3 is phosphorylated on Ser225 by ATR-kinase and this phosphorylation event is critical for RAD51 loading [41]. More recently, RAD51D protein levels were shown to be ubiquitin regulated by the E3-ubiquitin ligase, RNF138 [42, 43]. Having identified two RAD51D cancer-associated mutations in the N-terminus that influence its protein interactions and HR function, we wanted to explore the possibility that RAD51D might also be regulated through post-translational modifications in this region. To determine the potential role of RAD51D post-translational modifications, we collated mass spectrometry data on modified peptides from the publicly available database, PhosphoSitePlus (Table 3) [44]. We identified four residues in RAD51D that were modified by either phosphorylation or, in the case of lysine, acetylation (Tyr98, Thr99, Thr236 and K261R; Figure 2C). Residues Tyr98 and Thr99 were of particular interest as they resided between the Gly96 and Gly107 residues needed for RAD51D protein-protein interactions and HR (Figure 2C).
Table 3.
- RAD51D PTMs identified in the PhosphoSitePlus database
We then tested whether mutation of these residues disrupted the interaction of RAD51D with XRCC2 or RAD51C using the Y2H and Y3H assays, respectively. Residues were tested by either substituting an unmodifiable amino acid residue (mutation to alanine) or a phosphomimetic residue (mutation to glutamate). Secondly in the case of tyrosine, as there is not a suitable substitution to introduce a negative charge onto the benzene ring, only the unmodifiable substitution was introduced (mutation to Alanine or Phenylalanine in the double-mutant). Finally, the acetylated lysine was substituted with arginine, which cannot be modified. Additionally, as two of the residues are adjacent to each other (Tyr98 and Thr99), a double-mutant was generated to account for any potential redundancy between these phosphorylation sites. We find that none of the substitution mutations in RAD51D disrupted its Y2H or Y3H interaction with either XRCC2 or RAD51C (Figure 4A and B).
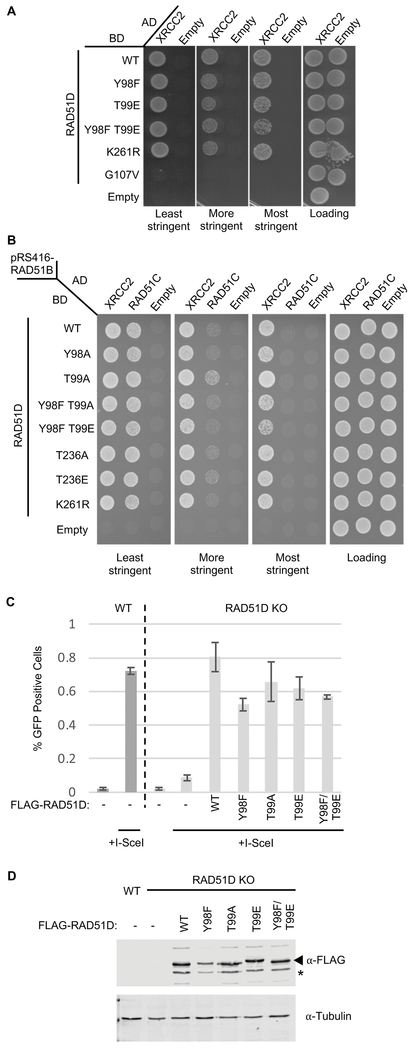
Figure 4. Mutations of residues in RAD51D that are potentially post-translationally modified do not affect the interaction of RAD51D with RAD51C or XRCC2 and do not affect HR-proficiency.
A) Yeast 2-hybrid analysis of potential post-translationally modified residues in RAD51D on its interaction with XRCC2. The PJ694a yeast strain was transformed with a plasmid where RAD51D or the indicated RAD51D mutation was fused to the GAL4-DNA binding domain [BD; pGBD-RAD51D (WT)] and a plasmid where XRCC2 was fused to the GAL4-DNA activating domain (AD; pGAD-XRCC2). A yeast-2-hybrid interaction was assayed by platting the yeast on SC-Leu¯Trp¯His¯ (least stringent), SC-Leu¯Trp¯His¯+3AT (more stringent), or SC-Leu¯Trp¯His¯Ade¯(most stringent) and compared to the loading control (SC-Leu¯Trp¯). Empty AD (pGAD; Empty) plasmid was used as a negative control.
B) Yeast 3-hybrid analysis of potential post-translationally modified residues in RAD51D on its interaction with XRCC2 and RAD51C. The PJ694a yeast strain was transformed with three plasmids; 1) a plasmid where RAD51D (or the indicated mutant) was fused to the GAL4-DNA binding domain [BD; pGBD-RAD51D (WT)], 2) a plasmid where either XRCC2 or RAD51C was fused to the GAL4-DNA activating domain (AD; pGAD-XRCC2, pGAD-RAD51C), and 3) a plasmid that constitutively expressed RAD51B (pRS416- RAD51B). Note that a plasmid containing RAD51B was used to help stabilize RAD51C expression. A yeast-3-hybrid interaction between RAD51D with XRCC2 or RAD51C was assayed by platting the yeast on SC-Leu¯Trp¯Ura¯His¯ (least stringent), SC-Leu¯Trp¯Ura¯His¯+3AT (more stringent), or SC-Leu¯Trp¯Ura¯His¯Ade¯(most stringent) and compared to the loading control SC-Leu¯Trp¯Ura¯ (Loading). Empty pGBD (BD Empty) and pGAD (AD Empty) plasmids were used as negative controls.
C) U20S wild-type (WT) or RAD51D knock-out cells were seeded on 35mm plates 24- hours prior to transfection with FLAG-RAD51D cDNA mutant constructs. Cells were cotransfected with either l-Scel and FLAG-RAD51D cDNAs containing residue substitutions or l-Scel and an empty vector control. Cells were incubated for an additional 48-hours after which, cells were analyzed by flow cytometry for green- fluorescence. The bar chart shows the average percentage of GFP+ cells over three experiments. Error bars show one standard deviation from the mean.
D) U20S wild-type (WT) or RAD51D knock-out cells were seeded on 35mm plates 24- hours prior to transfection with FLAG-RAD51D cDNA containing residue substitutions. Following transfection, cells were incubated for an additional 24-hours before lysis and Western blot analysis for RAD51D expression was done using αFLAG and αRAD51D antibodies. α-Tubulin was used as a loading control. The star indicates a non-specific band.
Although we find that substitution mutations that prevent or mimic protein modifications do not significantly alter any protein-protein interactions by Y2H or Y3H, it is possible that modification of these residues may be important for RAD51D HR function. To test whether substitution of residues Tyr98 and Thr99 impair RAD51D DNA repair function, we introduced RAD51D-Y98 and T99 mutations into RAD51D knock-out cells and determined whether they were expressed and HR proficient using a GFP-based sister chromatid recombination reporter assay (Figure 4C and 4D). Expression of both Y98A and T99A/E along with the double-mutant (T99F/T99E) as well as T236A/E and K261R successfully restored HR in RAD51D knock-out cells (Figure 4C; Supplemental Figure 1B). Therefore, modification of RAD51D residues, Y98, T99, T236 or K261 do not significantly alter RAD51D HR function.
Although we do not observe any HR-repair defects upon mutation of the post-translational modification sites in RAD51D, it is possible that these residues may be important for RAD51D function in other contexts such as replication-associated DNA damage. Further assays designed to test the functionality of RAD51D in response to different types of DNA damage or in response to different genotoxic agents will be necessary to test this.
3.5. Concluding Remarks
Here we identified isoform 1 of RAD51D to be the functional RAD51D isoform, and we identified two cancer-associated residues in the N-terminus (G96 and G107) that are independently important for RAD51D protein interactions and HR function. Both of these mutations are close to the Walker A motif, with G107V being the first glycine residue within the conserved Walker A motif. Previous studies have presented contradictory requirements for the Walker A and Walker B motifs for RAD51D function in response to DNA damage, specifically in tolerating DNA interstrand crosslinks. Gruver et al (2005) show that the RAD51D Walker A mutations K113A and K113R fail to restore resistance to DNA crosslinking agent, Mitomycin-C (MMC), in Rad5id-deficient mouse embryonic fibroblasts when compared to wild-type RAD51D [20], Furthermore, using the Y2H system, they demonstrate that RAD51D-K113A and K113R mutations primarily impair RAD51D’s interaction with RAD51C, while its interaction with XRCC2 was largely unaffected. In contrast, Wiese et al (2006) demonstrated that the same RAD51D mutations, K113A and K113R, maintain their interaction with both RAD51C and XRCC2 and that these mutants do indeed restore MMC resistance in Rad51d-knock-out CHO cells [22]. In addition, they showed that it is actually the RAD51D Walker B motif that is critical for its RAD51C and XRCC2 interactions and MMC resistance. By expressing RAD51D cDNAs with the same mutations (K113A/R) in RAD51D knock-out cell lines, we observe a partial requirement for the Walker A K113 residue for the repair of a DSB by HR. Although we cannot discount any cell line or organism-specific differences between these studies, our data supports the conclusion that conservation of the Walker A motif of RAD51D is required for both its protein-protein interactions with both RAD51C and XRCC2 as well as its HR function.
RAD51D is now included on the comprehensive breast and ovarian cancer screening panels [47, 48]. However, the vast majority of mutations identified remain variants of unknown significance. This has important clinical ramifications for the development of precision medicine approaches for HR-deficient tumors. Most notably, the use of poly-ADP-ribose polymerase inhibitors (PARPi) has revolutionized breast and ovarian cancer treatment [49–51]. PARPi exploit a synthetic lethality created by the loss of BRCA1 and BRCA2 mutations commonly observed in breast and ovarian cancer predisposition [52, 53]. BRCA1 and BRCA2 loss results in HR deficiency [54, 55], and this opens the possibility that other HR-deficient tumors will similarly respond to PARPi therapy. Therefore, the ability of cancer cells to repair DSBs by HR is key in predicting the sensitivity and response to PARPi. Previously, BRCA½ mutations were utilized to indicate HR-proficiency, however BRCA mutation alone does not accurately predict PARPi sensitivity in clinical trials [56]. By understanding which regions of RAD51D are important for its protein interactions and ultimately HR function, we will be better able to predict who might be a good candidate for PARPi therapy.
Supplementary Material
Highlights.
Only RAD51D isoform 1 rescues HR-deficiency observed in RAD51D knock-out cell lines
The RAD51D Walker A motif is important for its HR-function and interaction with XRCC2
Phosphorylation near the Walker A motif is unlikely to regulate RAD51D function
Acknowledgements
This work was supported by the National Institutes of Health [ES024872, GM088413 to K.A.B.], an American Cancer Society Research Scholar Grant [RSG-16-043-01-DMC to K.A.B], Stand Up to Cancer Innovative Research Grant [SU2C-AACR-IRG-02-16 to K.A.B.], and the V Foundation for Cancer Research V Scholar Award. This project used the Hillman Cytometry Facility that is supported in part by award P30CA047904. We would like to thank Paul Russell for providing us with the pCDNA3.1 FI_AG-RAD51D expression plasmid and Edwige B. Garcin and Stéphanie Gon for providing us with the RAD51D knock-out cell lines with the integrated sister chromatid recombination reporter assay. We also would like to thank Meghan Sullivan for providing invaluable feedback on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Jasin M and Rothstein R, Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol, 2013. 5(11): p. a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renkawitz J, Lademann CA, and Jentsch S, Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol, 2014. 15(6): p. 369–383. [DOI] [PubMed] [Google Scholar]
- 3.Chun J, Buechelmaier ES, and Powell SN, Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol, 2013. 33(2): p. 387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, et al. , hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem, 2011. 286(48): p. 41758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godin SK, Sullivan MR, and Bernstein KA, Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochemistry and Cell Biology, 2016: p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masson J-Y, et al. , Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes & Development, 2001. 15(24): p. 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JY, et al. , Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene, 2014. 33(40): p. 4803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somyajit K, et al. , Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res, 2015. 43(20): p. 9835–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarsounas M, et al. , Telomere maintenance requires the RAD51D recombination/repair protein. Cell, 2004. 117(3): p. 337–47. [DOI] [PubMed] [Google Scholar]
- 10.Kurumizaka H, et al. , Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem, 2002. 277(16): p. 14315–20. [DOI] [PubMed] [Google Scholar]
- 11.Godin SK, et al. , The Shu complex promotes error-free tolerance of alkylation-induced base excision repair products. Nucleic Acids Res, 2016. 44(17): p. 8199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krivokuca A, et al. , RAD51C mutation screening in high-risk patients from Serbian hereditary breast/ovarian cancer families. Cancer Biomark, 2015. 15(6): p. 775–81. [DOI] [PubMed] [Google Scholar]
- 13.Ollier M, et al. , DNA repair genes implicated in triple negative familial non-BRCA½ breast cancer predisposition. Am J Cancer Res, 2015. 5(7): p. 2113–26. [PMC free article] [PubMed] [Google Scholar]
- 14.Pelttari LM, et al. , RAD51, XRCC3, andXRCC2 mutation screening in Finnish breast cancer families. Springerplus, 2015. 4: p. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelttari LM, et al. , RAD51B in Familial Breast Cancer. PLoS One, 2016. 11(5): p. e0153788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, et al. , Contribution of Germline Mutations in the RAD51B, RAD51C, and RAD51D Genes to Ovarian Cancer in the Population. J Clin Oncol, 2015. 33(26): p. 2901–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tung N, et al. , Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J Clin Oncol, 2016. 34(13): p. 1460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reh WA, et al. , The homologous recombination protein RAD51Dprotects the genome from large deletions. Nucleic Acids Res, 2017. 45(4): p. 1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu Z, et al. , Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in ap53(−/−) background. Mol Cell Biol, 1999. 19(12): p. 8686–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruver AM, et al. , The ATPase motif in RAD51D is required for resistance to DNA interstrand crosslinking agents and interaction with RAD51C. Mutagenesis, 2005. 20(6): p. 433–40. [DOI] [PubMed] [Google Scholar]
- 21.O’Regan P, et al. , XRCC2 is a nuclear RAD51-likeprotein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem, 2001. 276(25): p. 22148–53. [DOI] [PubMed] [Google Scholar]
- 22.Wiese C, et al. , Disparate requirements for the Walker A andB ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res, 2006. 34(9): p. 2833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deans B, et al. , Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. The EMBO Journal, 2000. 19(24): p. 6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuznetsov SG, et al. , Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res, 2009. 69(3): p. 863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suwaki N, Klare K, and Tarsounas M, RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol, 2011. 22(8): p. 898–905. [DOI] [PubMed] [Google Scholar]
- 26.Puget N, Knowlton M, and Scully R, Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst), 2005. 4(2): p. 149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin V, et al. , Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J, 2006. 25(11): p. 2564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson C, Moynahan ME, and Jasin M, Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev, 1998. 12(24): p. 3831–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittman DL, Weinberg LR, and Schimenti JC, Identification, Characterization, and Genetic Mapping of Rad51d, a New Mouse andHumanRAD51/RecA-Related Gene. Genomics, 1998. 49(1): p. 103–111. [DOI] [PubMed] [Google Scholar]
- 30.Gruver AM, et al. , Functional characterization and identification of mouse Rad51d splice variants. BMC Mol Biol, 2009. 10: p. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couch FJ, et al. , Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol, 2017. 3(9): p. 1190–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller KA, et al. , Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Research, 2004. 32(1): p. 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schild D, et al. , Evidence for Simultaneous Protein Interactions between Human Rad51 Paralogs. Journal of Biological Chemistry, 2000. 275(22): p. 16443–16449. [DOI] [PubMed] [Google Scholar]
- 34.McClendon TB, et al. , Promotion of Homologous Recombination by SWS-1 in Complex with RAD-51 Paralogs in Caenorhabditis elegans. Genetics, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adzhubei IA, et al. , A method and server for predicting damaging missense mutations. Nat Methods, 2010. 7(4): p. 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherry ST, et al. , dbSNP: the NCBIdatabase of genetic variation. Nucleic Acids Res, 2001. 29(1): p. 308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson ER, et al. , Analysis of RAD51D in ovarian cancer patients and families with a history of ovarian or breast cancer. PLoS One, 2013. 8(1): p. e54772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George J, et al. , Comprehensive genomic profiles of small cell lung cancer. Nature, 2015. 524(7563): p. 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peifer M, et al. , Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas N, Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 2015. 517(7536): p. 576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somyajit K, et al. , ATM-andATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol Cell Biol, 2013. 33(9): p. 1830–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yard BD, et al. , RNF138 interacts with RAD51D and is required for DNA interstrand crosslink repair and maintaining chromosome integrity. DNA Repair, 2016. 42: p. 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han D, et al. , Ubiquitylation of Rad51dMediated by E3 Ligase Rnf138 Promotes the Homologous Recombination Repair Pathway. PLoS One, 2016. 11(5): p. e0155476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornbeck PV, et al. , PhosphoSitePlus: a comprehensive resource for investigating the structure and junction of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res, 2012. 40(Database issue): p. D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kettenbach AN, et al. , Quantitativephosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal, 2011. 4(179): p. rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merlins P, et al. , Proteogenomics connects somatic mutations to signalling in breast cancer. Nature, 2016. 534(7605): p. 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford B, et al. , Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res Treat, 2017. 163(2): p. 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Leary E, et al. , Expanded Gene Panel Use for Women With Breast Cancer: Identification and Intervention Beyond Breast Cancer Risk. Ann Surg Oncol, 2017. 24(10): p. 3060–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audeh MW, et al. , Oralpoly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: aproof-of-concept trial. Lancet, 2010. 376(9737): p. 245–51. [DOI] [PubMed] [Google Scholar]
- 50.Tutt A, et al. , Oralpoly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet, 2010. 376(9737): p. 235–44. [DOI] [PubMed] [Google Scholar]
- 51.Gelmon KA, et al. , Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomisedstudy. Lancet Oncol, 2011. 12(9): p. 852–61. [DOI] [PubMed] [Google Scholar]
- 52.Bryant HE, et al. , Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose)polymerase. Nature, 2005. 434(7035): p. 913–7. [DOI] [PubMed] [Google Scholar]
- 53.Farmer H, et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature, 2005. 434(7035): p. 917–21. [DOI] [PubMed] [Google Scholar]
- 54.Xia F, et al. , Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A, 2001. 98(15): p. 8644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moynahan ME, et al. , Brca1 controls homology-directed DNA repair. Mol Cell, 1999. 4(4): p. 511–8. [DOI] [PubMed] [Google Scholar]
- 56.Swisher EM, et al. , Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. The Lancet Oncology, 2017. 18(1): p. 75–87. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, et al. , The yeast Shu complex utilizes homologous recombination machinery for error-free lesion bypass via physical interaction with a Rad51 paralogue. PLoS One, 2013. 8(12): p. e81371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.