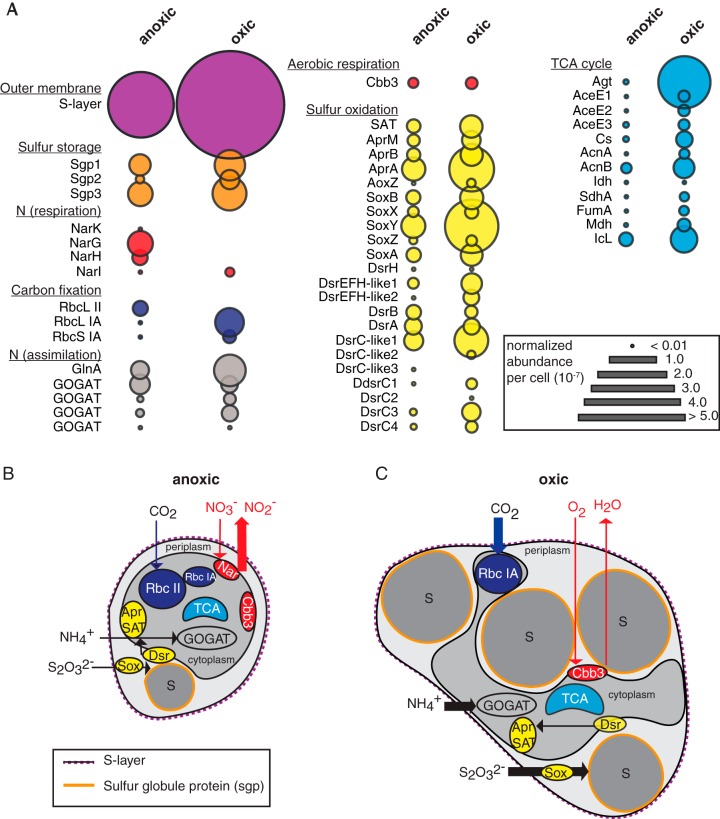
FIG 4.
Protein expression in “Ca. Thioglobus autotrophicus” cells under anoxic and oxic growth conditions in thiosulfate-replete media. (A) Normalized relative protein abundance per cell of the S-layer protein and other key proteins involved in carbon, nitrogen, and sulfur metabolisms under anoxic and oxic growth conditions. (B and C) Schematic of “Ca. Thioglobus autotrophicus” cell structure and metabolism under anoxic (B) and oxic (C) growth conditions. Schematics are based on cell size, CHN content, and protein expression-level data. Major processes are color coded as in panel A for carbon, nitrogen, and sulfur metabolic pathways. Arrows indicate direction and magnitude of major nutrient fluxes. Sgp, sulfur globule protein; Nar, dissimilatory nitrate reductase; Rbc, ribulose-1,5-bisphosphate carboxylase/oxygenase; Gln, glutamine synthetase; GOGAT, glutamine oxoglutarate aminotransferase; Cbb3, cytochrome c oxidase (Cox); SAT, ATP sulfurylase; Apr, adenosine phosphosulfate reductase; Sox, sulfur oxidation system; Dsr, dissimilatory sulfite reductase system. Additional information about the identified proteins is available in Dataset S1.