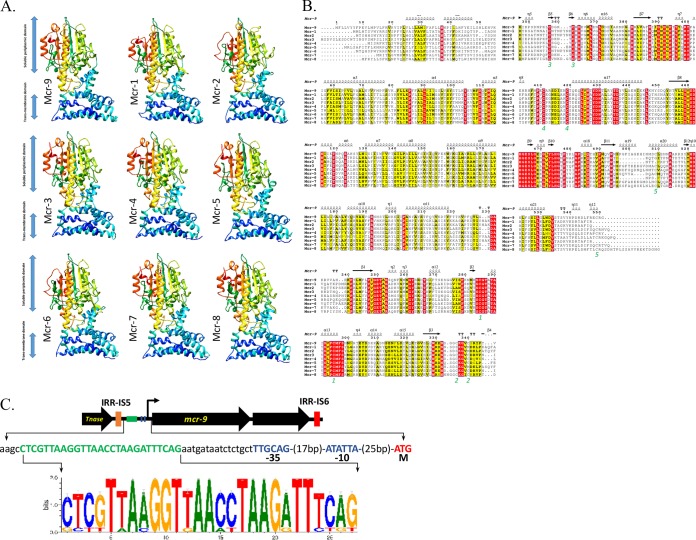
FIG 2.
(A) Structural models of all published Mcr proteins (Mcr-1 to -8) and Mcr-9, based on lipooligosaccharide phosphoethanolamine transferase EptA. Models were constructed using the Phyre2 server, and structures were viewed and edited using UCSF Chimera. Structural models show conservation of two EptA domains: transmembrane-anchored and soluble periplasmic domains. (B) Location of Mcr-9 secondary structure elements within the alignment of Mcr amino acid sequences, constructed using the ESPript 3 server. The top track denotes Mcr-9 secondary structure elements (alpha helixes and beta sheets). Green digits below the alignment denote cysteine residues forming a disulfide bridge (e.g., 1 forms a bridge with 1, 2 with 2, etc.). Within the amino acid sequence alignment itself, a strict identity (i.e., identical amino acid residue at a site) is denoted by a red box and a white character. A yellow box around an amino acid residue denotes similarity across groups, where groups were defined using the default “all” specification in ESPript 3 (ESPript 3 total score [TSc] > in-group threshold [ThIn]), while a residue in boldface denotes similarity within a group (ESPript 3 in-group score [ISc] > ThIn). (C) Organization of the mcr-9 locus in S. Typhimurium. An unknown function cupin fold metalloprotein is encoded by the gene downstream of mcr-9 (unlabeled black arrow). The mcr-9 locus is flanked by two different terminal repeat sequences (IRR) from the IS5 (orange box) and IS6 (red box) families. The mcr-9 upstream region contains highly conserved putative −35 and −10 σ70-dependent promoter elements (blue boxes and blue text). Moreover, the mcr-9 promoter region contains an inverted repeat motif (green box, green text, and sequence logo) that is conserved in more than 95% of 321 mcr-9 genes, as shown by the sequence logo (constructed using WebLogo) (24).