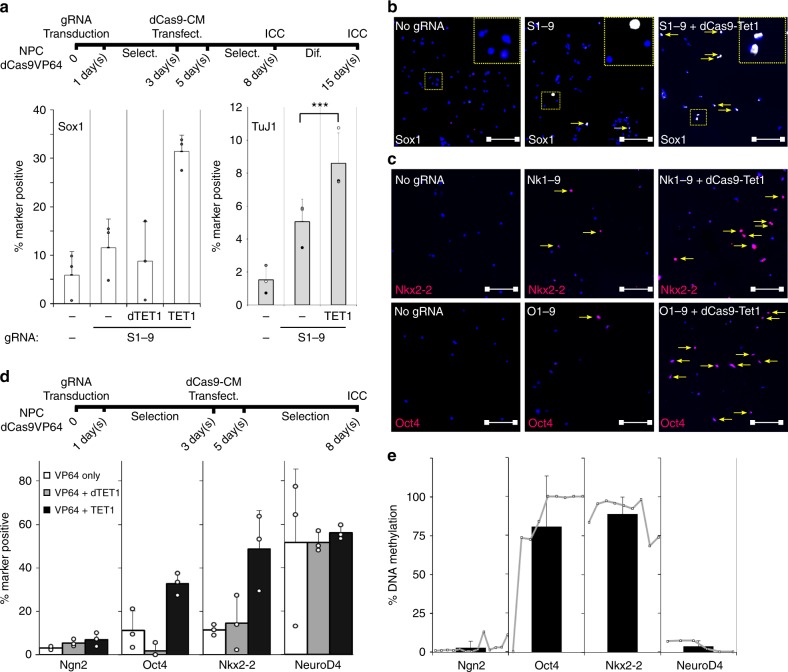
Fig. 6.
DNA methylation as a barrier to transcriptional engineering is not an exclusive feature of Sox1. a, b Combination of transcriptional engineering with epigenome editing increases the neuronal differentiation potency of NPCs. NPCs stably expressing dCas9-VP64 were transduced with Sox1-targeting gRNAs (S1-9) and subsequently transfected with dCas9-Tet1 or dCas9-dTet1. NPCs were stained for Sox1 or differentiated for 7 more days, respectively. Sox1 staining revealed a higher number of Sox1-positive cells when dCas9-VP64 is combined with dCas9-Tet1 but not with dCas9-dTet1. After differentiation, more cells differentiated into the neuronal lineage, as indicated by the higher Tuj1 positivity after epigenome editing compared to transcriptional editing alone. Data are shown as the mean and standard error of the mean of n = 3 biological replicates, performed on different days in different clonal lines. ***p < 0.005 calculated by two-sided Student’s t-test. Dashed yellow lines mark magnified areas; scale bar: 100 µm. c, d Activation of different master transcription factors reveals varying responsiveness to transcriptional engineering and sensitivity to DNA demethylation. NPCs expressing dCas9-VP64 were transfected with gRNAs (Ng1-9, Nk1-9, O1-9, Ne1-9) targeting different master transcription and reprogramming factors (Ngn2, Nkx2-2, Oct4, NeuroD4). Induction varied considerably between different targets, although three of four master transcription factor genes mostly resisted gene activation. Transfection of dCas9-Tet1 but not dCas9-dTet1 almost quintupled the cell population responsive to Nkx2-2 transactivation and tripled that inducing Oct4. Data shown as the mean with standard error of the mean of n = 3 technical replicates; scale bar: 100 µm. e DNA methylation levels at master transcription factor promoters predict responsiveness to epigenome editing. Oxidative bisulfite sequencing was performed in NPCs to quantify DNA methylation levels at regions including the TSS of the tested master transcription factors. While the promoter of Ngn2 and NeuroD4 appear almost completely unmethylated, that of Nkx2-2 and Oct4 exhibit high methylation levels at the TSS. Data derived from two biological replicates are shown as the mean and standard error of the mean of all analyzed CpGs (Ngn2: 13, Nkx2-2: 8, Oct4: 9, NeuroD4: 6) inside a 100–300 bp region surrounding the TSS (for the genomic position see Supplementary Table 1); Dots show the methylation levels of single CpGs in analyzed loci