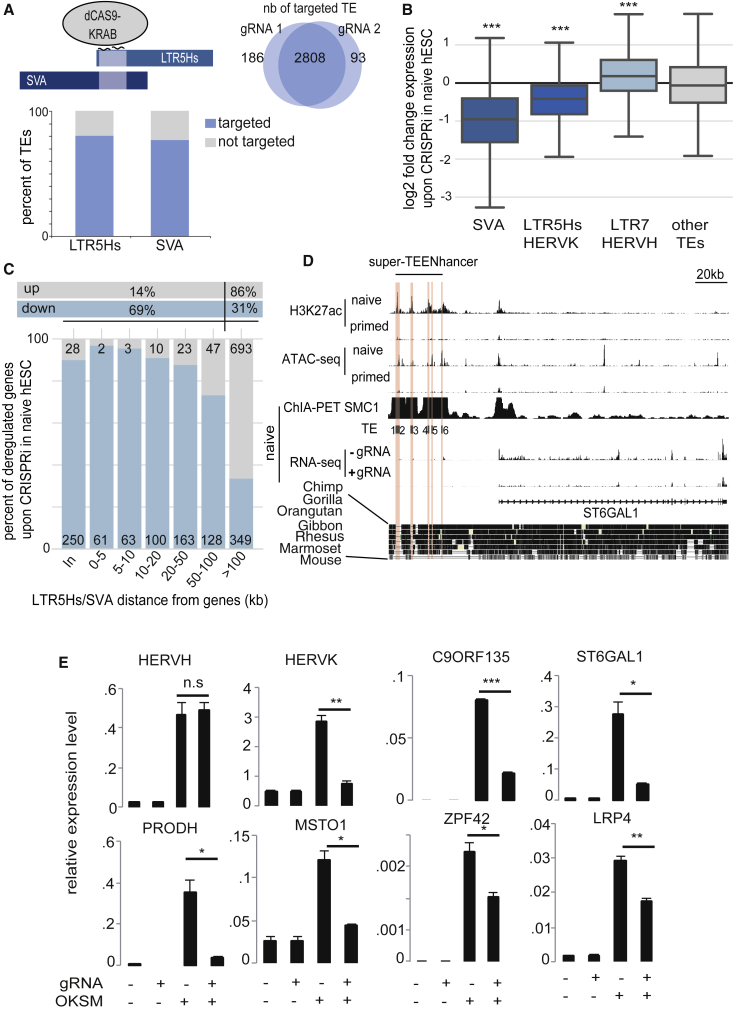
Figure 3.
TEENhancers Regulate Gene Expression in Naive hESCs
(A) Schematic representation of CRISPRi targeting of SVA-LTR5Hs common region. Venn diagram depicts the number of TEs predicted to be targeted by either gRNA, with bar plot indicating the percentage of the 2,808 commonly targeted integrants within each subfamily.
(B) Impact of CRISPRi on TEs expression. Naive hESCs were transduced with a dCAS9-KRAB lentiviral vector containing or not gRNAs against LTR5Hs-SVA (two different gRNAs each in quadruplicate). Expression of indicated TEs was compared between gRNA-expressing and control cells. One-sample t tests were computed to generate p values. ∗∗∗p ≤ 0.001.
(C) Impact of TE-targeting CRISPRi on gene expression. Number of up- and downregulated genes (p < 0.05 and fold change >10% between paired replicates) at indicated distance from closest CRISPRi-targeted TE is shown (in: TE within gene). (Top) Percentage of all up- or downregulated genes within 100 kb or farther away is shown.
(D) Long-range regulation of a gene by an early embryonic super-TEENhancer. (Top) H3K27ac, ATAC-seq, and ChIA-PET (Ji et al., 2016) profiles in naive and primed hESCs are shown. (Middle) RNA-seq in naive cells sorted for GFP-naive reporter in dCAS9-KRAB-transduced naive hESCs expressing (+) or not (−) the LTR5HS-SVA-targeting gRNA. (Bottom) Alignment of genomes of indicated species at homologous locus, revealing the human specificity of the TEENhancers. Then, TEs 1 through 6 (3 LTR5Hs and 3 SVAs) predicted to bind the gRNA are shaded in red.
(E) CRISPRi inhibits SVA-LTR5Hs-controlled gene activation during somatic reprogramming. Human primary fibroblasts containing dCAS9-KRAB with or without gRNA against LTR5Hs-SVA (gRNA+/−) were transduced or not (OSKM+/−) with OSKM-expressing Sendai virus for 6 days before measuring transcripts of HERVH, HERVK, and several genes found to interact with SVA-LTR5Hs by ChIA-PET and downregulated by CRISPRi in naive cells (normalized to RPLP0). Error bars represent SEM while the p value was established with a t test (∗∗∗ ≤ 0.001, ∗∗ ≤ 0.01, ∗ ≤ 0.01, and n.s. > 0.05).