Figure 4.
Evolutionary Recent KZFPs Tame TEENhancers during Human Early Embryogenesis
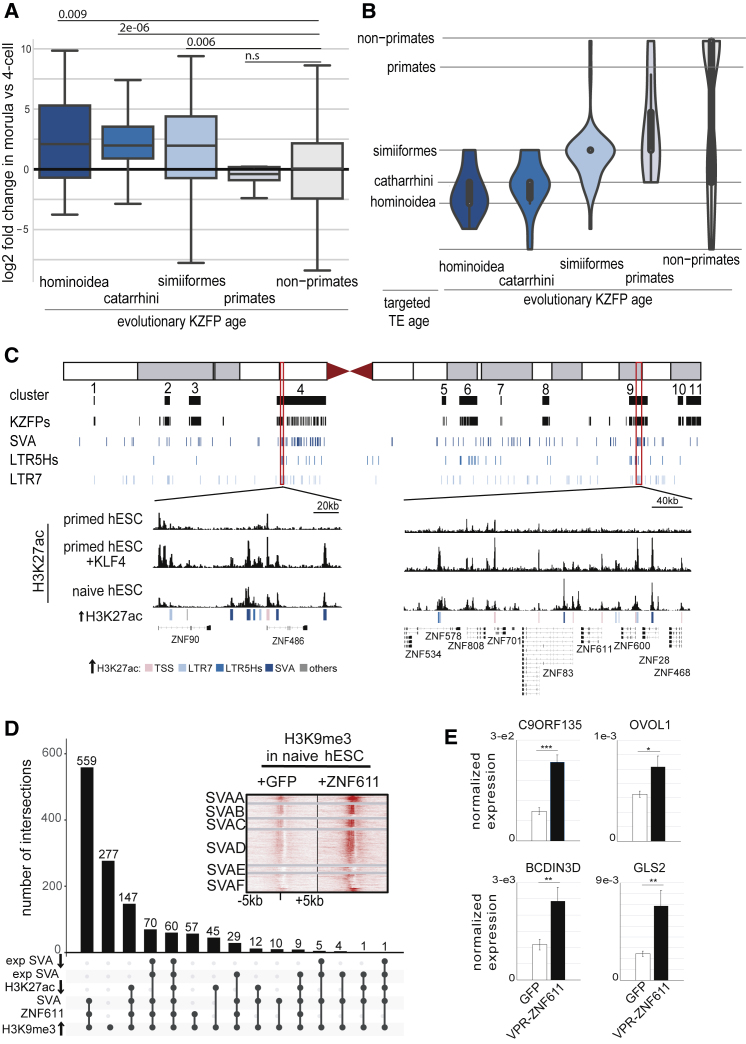
(A) Evolutionary young KZFPs are activated during EGA. Fold change expression of KZFP genes (classified by evolutionary age; Imbeault et al., 2017) between 4C and morula (data re-analyzed from Yan et al., 2013).
(B) Young KZFPs target contemporaneous TEs. Violin representation of evolutionary ages of KZFPs and their TE targets, determined by re-analyzing ChIP-exo data (Imbeault et al., 2017) and using the most significantly bound TE for each EGA-induced KZFP. TEs were classified by evolutionary age established through lineages comparison.
(C) KLF4 induces histone acetylation within evolutionary young KZFP clusters. (Top) Schematic representation of human chromosome 19 with KZFP gene clusters, individual units, and TEs distribution below are shown. (Bottom) Highlight of the H3K27ac profile of two regions in naive or primed hESCs with or without overexpression of KLF4 is shown. Underlying genetic entities are indicated by colored boxes.
(D) UpSet plot showing impact of ZNF611 overexpression in naive hESCs. Black dots crossed by black lines are the different combinations of intersections and are mutually exclusive of each other. Intersection combinations were plotted as bar chart representing the number of loci with increased H3K9me3 signal in naive hESCs overexpressing ZNF611 compared to GFP, split into subcategories according to whether they were known ZNF611 binding sites in 293T cells (ZNF611), SVAs (SVA), H3K27ac-depleted (H3K27ac ↓), expressed SVAs (exp SVA), and downregulated SVAs (exp SVA ↓). Heatmap illustrates H3K9me3 raw signal ±5 kb around all SVAs in naive hESCs overexpressing ZNF611 or GFP.
(E) Impact of activation domain fused to ZNF611 zinc fingers in primed hESCs. The VPR activation domain (comprising the corresponding regions of VP64, P65, and Rta) was fused to the zinc finger domain of ZNF611 and overexpressed in primed hESCs. Bars represent qRT-PCR expression levels (normalized to GAPDH). Error bars were established using SEM and p value using t test (∗∗∗ ≤ 0.001; ∗∗ ≤ 0.01; ∗ ≤ 0.05). Represented genes were selected on the following criteria: downregulated by ZNF611 overexpression in naive hESC; devoid of SVA inside of their gene body; and having a SVA enhancer within 3–30 kb of their TSS.
See also Figures S4 and S5.