FIGURE 2.
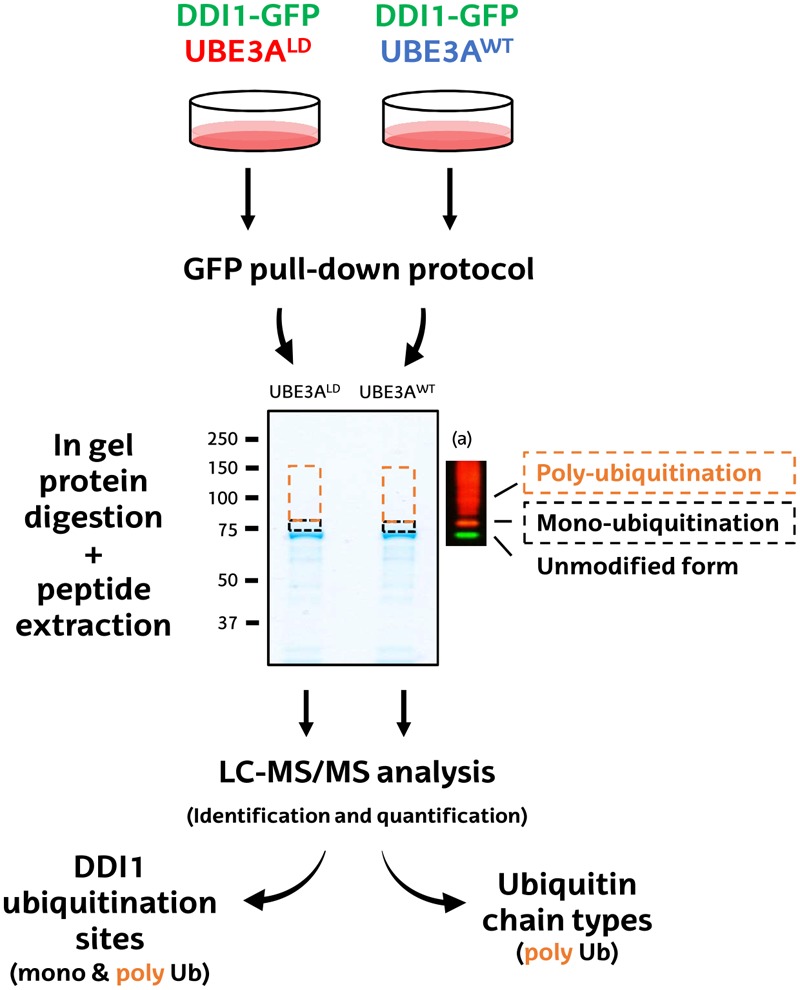
Schematic diagram of the MS-based procedure used to study ubiquitination sites and ubiquitin chain types on DDI1. HEK293T cells were co-transfected with DDI1-GFP and wild type UBE3A (UBE3AWT) or ligase dead UBE3A (UBE3ALD). Unmodified DDI1-GFP as well as its ubiquitinated forms were isolated by GFP pull-down, and separated by SDS–PAGE. The gel slice corresponding to ubiquitinated DDI1-GFP was excised and divided into two, basing on a previous western blot (a); a lower band corresponding for mono-ubiquitinated DDI1-GFP (black), and a band above it corresponding to poly-ubiquitinated DDI1-GFP (orange). Gel slices were digested with trypsin, and resulting peptides were extracted from the gel to further identify and quantify them by LC-MS/MS. Both bands were analyzed to infer DDI1 ubiquitination sites, whereas only the poly-ubiquitination band was used to detect the nature of the ubiquitin chain types formed on DDI1.
