Fig. 2.
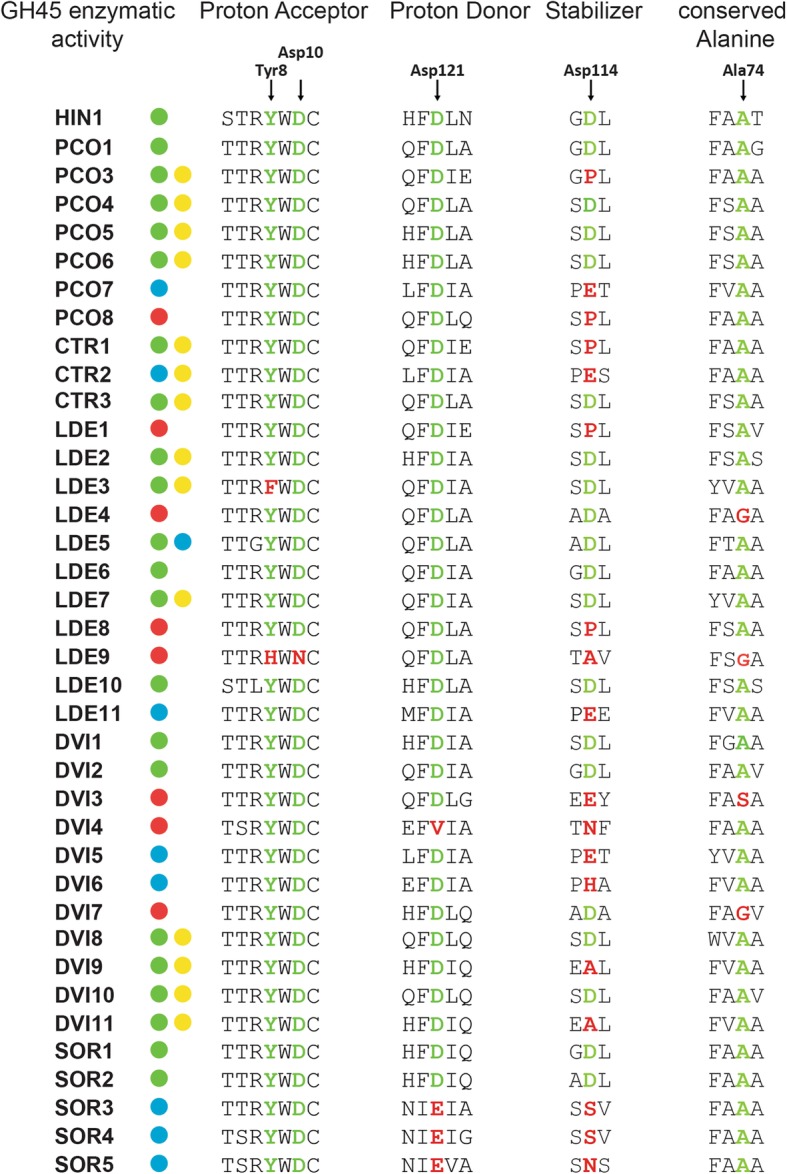
GH45 amino acid alignment of the catalytic residues. We used a GH45 sequence of Humicola insulens (HIN1) as a reference sequence (Accession: 2ENG_A) [37]. According to HIN1, we chose to investigate the catalytic residues (ASP10 and ASP121) as well as a conserved tyrosine (TYR8) of the catalytic binding site, a crucial substrate-stabilizing amino acid (ASP114) and an essential conserved alanine (ALA74). Arrows indicate amino acid residues under investigation. If the respective amino acid residue is highlighted in green, it is retained in comparison to the reference sequence; otherwise it is highlighted in red. GH45 enzymatic activity was color-coded based on the respective substrate specificity (green dots = endo-β-1,4-glucanase, blue dots = endo-β-1,4-xyloglucanase, yellow dots = (gluco)mannanase, red dots = no detected activity)
