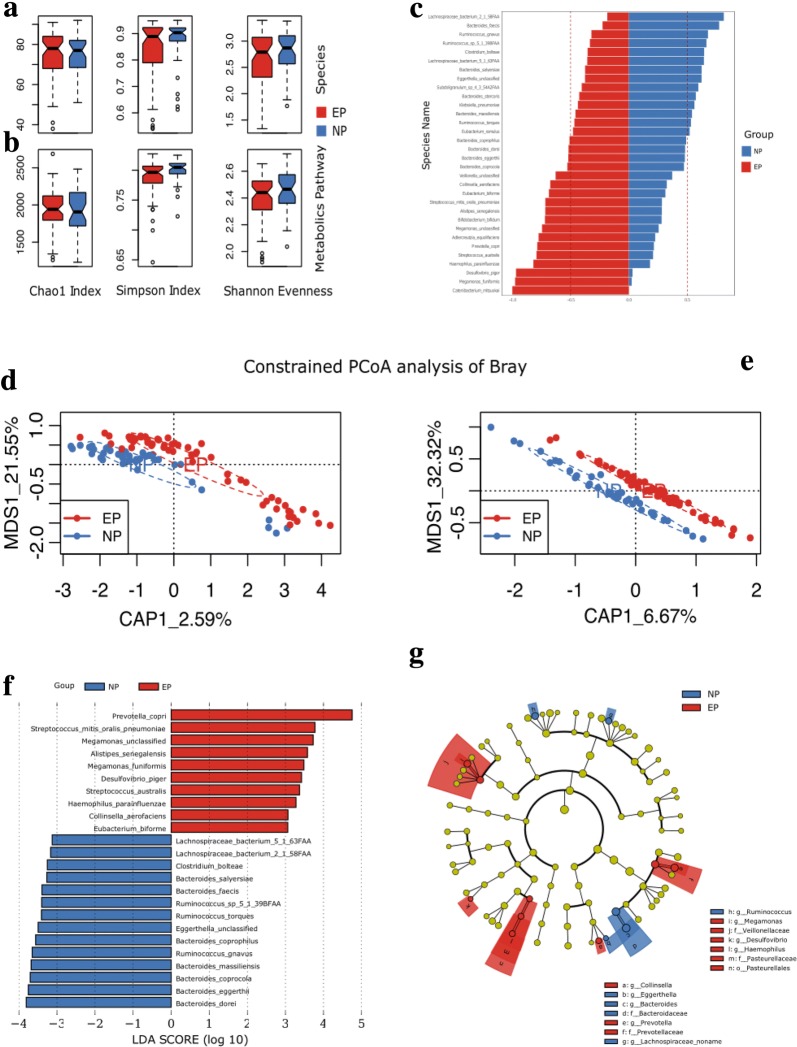
Fig. 1.
Microbiome and metagenomics diversity and LEfSe analysis. a, b Alfa-diversities of microbiome composition and functional capacities between EP (red) and NP (blue) groups. c Species that are significantly different in EP (red) vs. NP (blue) groups. Significance was determined using Wilcoxon rank-sum test, and the relative proportion is shown for each species. d, e Bray–Curtis based constrained principal co-ordinates analysis (PCoA) showing EP (red) and NP (blue) with significantly different taxonomical compositions and functional capacities. f, g The biomarkers identified by linear discriminant analysis effect size (LEfSe) ranked according to the effect size and associating them with the class with the highest median. The color red represents the biomarkers in the EP group and the blue color indicates biomarkers in the NP group. The length of each bar represents the linear discriminant analysis (LDA) score format with log 10