Abstract
Alaska Native and American Indian (AN/AI) people have unique pharmacogene variation that may affect warfarin disposition and therapeutic response. We performed targeted genotyping for cytochrome P450 (CYP)2C9, vitamin K epoxide oxidase reductase complex subunit 1 (VKORC1), CYP4F2,CYP4F11, and gamma‐glutamyl carboxylase (GGCX) variants in AN/AI people receiving warfarin. The primary outcome was stable warfarin dose, defined as one dose, and associated international normalized ratio within the target range, at least 6 months after starting therapy, with two matching doses at least 2 weeks apart. Genotype–phenotype relationships were assessed by multivariate regression analysis, adjusted for self‐reported heritage, age, gender, and concurrent statin use. VKORC1 genotype explained 34% of dose variability, with VKORC1 −1639G>A and 1173C>T associated with a 1.7 mg/day (P = 1.4e‐05) dose reduction. Additionally, CYP2C9 N218I was suggestively significant (P = 0.077), with heterozygotes requiring 1.1 mg/day less than reference individuals. Self‐reported heritage was significantly associated with dose, largely driven by differences in the diagnostic VKORC1 allele frequencies among AN/AI people.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ VKORC1, CYP2C9, GGCX, and CYP4F2 are pharmacogene‐encoding enzymes that have been associated with altered warfarin disposition and pharmacological response, but none of these genes have been studied in warfarin‐treated patients in the AN/AI community, challenging implementation of pharmacogenetic‐guided warfarin dosing in this population.
what question did this study address?
☑ This study investigated whether inheritance of known and novel variants in CYP2C9, VKORC1, GGCX, CYP4F2, and CYP4F11 affects the dose of warfarin required to achieve a therapeutic INR in the AN/AI population.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ VKORC1 genotype explains a higher percent of warfarin dose variability in the AN/AI population than that observed in other world populations, and the novel CYP2C9 N218I coding variant meaningfully lowers warfarin dose requirement.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Prospective pharmacogenetic screening for VKORC1 and CYP2C9 variation could help guide initial dose selection to improve the warfarin safety and efficacy in this underserved population.
The oral vitamin K antagonist warfarin (Bristol‐Myers Squibb Company, Princeton, NJ) is used to prevent stroke in patients with atrial fibrillation and for secondary prevention of venous thromboembolism.1 Despite newer treatment options, such as the direct oral anticoagulants, warfarin remains a mainstay in anticoagulation therapy and is the most frequently prescribed anticoagulant in the United States.2 Warfarin therapy requires intensive monitoring and dose titration due to its narrow therapeutic index and wide interindividual (up to 30‐fold) response, due in part to genetic variation,3 as well as clinical, demographic, and environmental factors.4
Although variation in vitamin K oxidoreductase complex 1 (VKORC1), cytochrome P450 (CYP)2C9, CYP4F2, and gamma‐glutamyl carboxylase (GGCX) genes have all been associated with the warfarin dose needed to achieve a common degree of anticoagulation response,5, 6, 7, 8, 9 these findings are based largely on alleles and associated frequencies discovered in populations with limited ethnic/racial heterogeneity (i.e., white, Asian, and African American) and may not be generalizable to other, less studied populations, such as the indigenous people of North America.10, 11 In that regard, we recently reported the discovery within the indigenous population of Alaska (Alaska Native and American Indian (AN/AI)) of common, novel coding variation in the CYP2C9 gene (M1L and N218I) predicted to decrease CYP2C9 enzyme function.12 We also observed relatively high frequencies of known function‐altering VKORC1 (−1639G>A) and CYP4F2 (*3, V433M) alleles in some but not all AN/AI subgroups. These data warrant further investigation to understand their clinical impact on warfarin therapy in AN/AI people. Variation in CYP4F11 was included for testing, as the gene product can also catalyze vitamin K catabolism.13 Thus, the goal of this project was to determine whether inheritance of CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX gene variants, particularly novel variants in an AN/AI population, affect the dose of warfarin required to achieve a therapeutic international normalized ratio (INR) in order to better understand the significance of genetic testing to guide warfarin therapy for the AN/AI population and potentially other indigenous peoples of North America.
Methods
Setting
The Southcentral Foundation (SCF), a tribally owned and operated regional health corporation, provides prepaid healthcare services to 65,000 AN/AI customer‐owners. The Anchorage Service Unit and Cook Inlet Region Villages served by the SCF are comprised of both urban and rural areas, including Anchorage, the Matanuska‐Susitna Borough, and 76 outlying villages (most with fewer than 500 residents). It provides primary care services to 46% of the AN population in the Anchorage Service Unit at six SCF primary care clinics on the Alaska Native Medical Center campus where participant recruitment took place.
Study participants
Between 2011 and 2013, a representative convenience sample of 118 AN/AI customer‐owners, ≥ 18 years of age, receiving warfarin therapy at SCF, were recruited, and consent obtained by research staff members at SCF's primary care clinics. Study participants completed a short demographic questionnaire (self‐reported gender, date of birth, and self‐reported heritage). Consented customer‐owners were then provided two small, sterile swabs to collect epithelial cheek cells for DNA analysis of CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX gene variations. Swabs were then placed in sterile tubes and were stored at −80°C until genotyping analysis.
Study design
The Alaska Area institutional review board and the SCF and Alaska Native Tribal Health Consortium tribal review boards approved work conducted at SCF on the Alaska Native Medical Center campus. The University of Washington institutional review board approved the overall research project, as University of Washington is the academic home of the grant funding this research (Pharmacogenetics in Rural and Underserved Populations) and its principal investigators. The National Institute of General Medical Sciences and the Indian Health Service granted a Certificate of Confidentiality for protection of customer‐owner information, and the respective Alaska Area institutional review board approved forms for written consent prior to initiating research. Community‐based participatory research at SCF and the Center for Alaska Native Health Research were used to develop research questions.
This retrospective cohort study was conducted at one anticoagulation clinic based in Anchorage, Alaska. Customer‐owner care for this study was managed by a credentialed anticoagulation pharmacist with physician oversight. A standardized approach aided by commercial anticoagulation software was used, and follow‐up averaged a little more than 2 weeks. All customer‐owners < 65 years of age received the same initial dose, 5 mg/day, with subsequent dose adjustments made based on INR results.
Customer‐owners’ medical records were retroactively queried by the SCF Data Services Department staff for specific data elements (i.e., International Classification of Diseases, Ninth Revision, codes and diagnoses, purpose, and explanation of the data queried). For elements extracted prior to October 1, 2011, the Legacy electronic medical record system Resource Patient Management System was used, and for dates on or later than October 1, 2011, the Cerner electronic medical record system was used. Data abstracted included dates of warfarin initiation/stabilization and indication for treatment (deep vein thrombosis, pulmonary embolism, pulmonary hypertension, atrial fibrillation/flutter, cardiomyopathy, left ventricular dilation, stroke, postorthopedic variables, valve replacement, bleeding events, and thromboembolic events). Comorbidities recorded included chronic liver disease, heart disease, diabetes, valve replacement, stroke, anemia, kidney dysfunction, ulcers, hyperlipidemia, and cancer, as well as the use of medications known to interact with warfarin. The following demographic/background variables that may impact dose of warfarin or likelihood of bleeding events were also collected: gender, age, heritage, height, and weight. Password‐protected electronic databases were used to store customer‐owner questionnaire and medical record information.
To account for population substructure within the SCF cohort, customer‐owners were asked for self‐reported tribal affiliation. Customer‐owners classified as AN included the following tribes: Inupiaq, Athabascan subgroups, Tlingit, Tsimshian, Haida, Eyak, Aleut/Unangan, Central Yup'ik, Cup'ik, and Sugpiaq/Alutiiq. Customer‐owners also were given the option of choosing affiliation with multiple tribes in the lower 48 states of the United States and were classified as AI. Due to the small sample size, we intentionally grouped these customer‐owners by their shared heritage and geographic proximity for data analysis, although we acknowledge that each tribe has its own unique culture and history.
Genotyping methods and linkage disequilibrium calculations
Genomic DNA from buccal swabs was extracted using a QIAamp DNA Blood Midi/Maxi kit (Qiagen, Valencia, CA). Quality and concentration of DNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Population‐relevant single nucleotide variations (SNVs) and small insertions/deletions were identified previously through gene resequencing in 188 AN/AI customer‐owners in partnership with SCF in Anchorage, Alaska.12 DNA samples from the current study participants were genotyped for the novel and known variants of interest using the Fluidigm Platform (South San Francisco, CA), as previously reported.12 This included eight SNVs in CYP2C9, four in CYP4F2, one in CYP4F11, two in VKORC1, and two in GGCX. Samples with overall call rates below 95% were removed from further analysis. Of all DNA samples genotyped, nine were excluded from further analysis due to call rates below 95%. The no‐call rate was 0.4%. Allele frequencies and pairwise linkage disequilibrium were calculated using Haploview version 4.2 software.14 All SNVs identified were tested for deviations from Hardy–Weinberg equilibrium using a χ2 test. Allele frequency results were compared with previously reported AN/AI sequencing results from SCF customer‐owners.12
Outcomes
The outcome variable for this study was stable therapeutic warfarin dose (mg/day), which was ascertained from available clinical data and prescription records from January 2000 to June 2017. Stable warfarin dose was defined in two ways: (INR‐based) the dose of warfarin required to achieve an INR within the target range, at least 6 months after starting warfarin therapy, with two matching doses, at least 2 weeks apart; or (consecutive, non‐INR‐based) two matching consecutive doses at least 6 months after starting warfarin therapy and at least 2 weeks apart. The INR was considered in therapeutic range if it was measured to be within the target range for a given indication. For example, if the target INR was 2.0–3.0, then the first INR result between 2.0 and 3.0, and at least 6 months after initiating therapy, was defined as within range. Once the customer‐owner was determined to reach their stable warfarin dose, his/her maintenance dose was recorded, allowing for comparison by various genotypes in warfarin pharmacogenes.
Statistical analysis
Customer‐owners were included in two groups based on available clinical data: INR‐based stable warfarin dose and consecutive (non‐INR‐based) dose. All customer‐owners included in the INR‐based cohort were also included in the consecutive cohort. For each cohort, associations between pharmacogenetics and pharmacodynamics were evaluated with a univariate and multivariate linear regression models using RStudio version 1.0.143 (RStudio, Boston, MA). To account for heteroscedasticity, robust SEs were calculated using the lmtest package in R. Covariates included in the International Warfarin Pharmacogenetics Consortium pharmacogenetic dosing algorithm were considered; however, no customer‐owners in this analysis were using CYP2C9 inducers concurrently and the races in the International Warfarin Pharmacogenetics Consortium algorithm (Asian, black, or white) were not used,15 and instead we used heritage (AN or AI). Height and weight were collected at the time of genotyping; however, this may not correspond to the same height and weight that a customer‐owner was at the time of reaching stable warfarin dose, so body mass index (BMI) was not included in the multivariate regression model.
We elected to include age, gender, and heritage in the multivariate regression model a priori, as these demographic variables have been shown to influence warfarin dose across many world populations.15 The potential confounding effects of the concurrent use of CYP2C9 inhibitors (amiodarone, fluconazole, and disulfiram), statins (simvastatin), or antibiotics on warfarin dose were also evaluated. Concurrent medication classes were added to the model one at a time to assess potential confounding effects on warfarin dose and were included in the multivariate regression model if the medication was used by at least 10 customer‐owners in the study population and if the covariate significantly impacted warfarin dose. The primary analysis assessed associations between single SNVs and INR‐based stable warfarin dose adjusting for age, gender, AN or AI heritage, and concurrent statin use in a multivariate regression model. For SNVs that included reference, heterozygote, and homozygous variant observations, an additive model was used for genotype.
Secondary analysis of the data was conducted, testing for associations with a combination of impaired function CYP2C9 variants as well as adjusting for SNVs found to be significantly associated with stable warfarin dose in the primary analysis model, allowing for detection of stronger, independent signals from other variants potentially associated with dose. The primary and secondary analyses were repeated in the consecutive (non‐INR‐based) stable warfarin dose cohort. Bonferroni adjusted P values were used to correct for multiple testing of five independent tests of the five candidate loci of interest. The SCF team independently validated the statistical analysis by reviewing and running all R code. Any discrepancies in interpretation were discussed among the team and consensus was reached.
Results
A total of 118 AN/AI customer‐owners at SCF receiving warfarin therapy were enrolled in this study. Figure 1 depicts the number of customer‐owners included in the final analysis, and Table 1 describes study participant characteristics and stable warfarin dose. The primary INR‐based stable warfarin dose definition (dose required to achieve an INR within the target range, at least 6 months after starting warfarin therapy, with two matching doses, at least 2 weeks apart) included 50 customer‐owners in the genotype–phenotype analysis, with 43 AN and 7 AI individuals. The secondary non‐INR‐based warfarin dose definition (two matching consecutive doses 6 months after starting warfarin therapy, and at least 2 weeks apart) included 78 customer‐owners, with 68 AN and 10 AI individuals. The average stable warfarin dose (5.0 vs. 4.9 mg/day) and range (1.7–13.0 vs. 1.5–12.5 mg/day) were similar between the INR‐based dose definition and the consecutive (non‐INR‐based) dose definition cohorts (Table 1).
Figure 1.
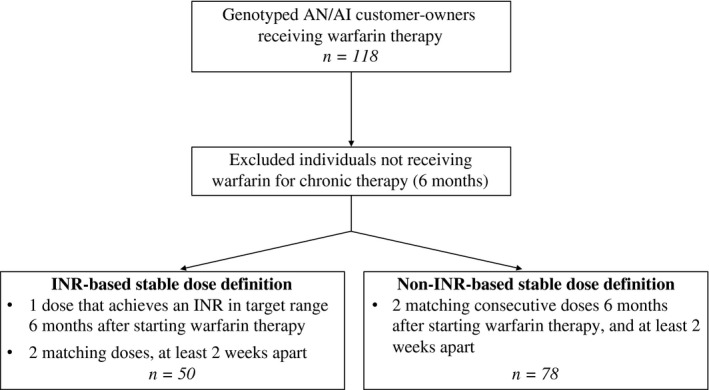
Flow diagram depicting the number of customer‐owners included and excluded in this study and the primary (international normalized ratio (INR)‐based) and consecutive (non‐INR‐based) stable warfarin dose definition cohorts. AI, American Indian; AN, Alaska Native.
Table 1.
Comparison of customer‐owner characteristics and warfarin dose in the INR‐based and consecutive (non‐INR‐based) stable warfarin dose definition cohorts
| Variable | INR‐based stable dose cohort (n = 50) | Consecutive stable dose cohort (n = 78) |
|---|---|---|
| Demographics | ||
| Alaska Native, no. (%) | 43 (86.0) | 68 (87.2) |
| American Indian, no. (%) | 7 (14.0) | 10 (12.8) |
| Men, no. (%) | 29 (58.0) | 45 (57.7) |
| Age, mean (SD), years | 57.9 (14.9) | 59.6 (15.0) |
| BMI, mean (SD) | 30.6 (7.6) | 30.7 (6.9) |
| Indication for warfarin, no. (%) | ||
| Cardiomyopathy/left ventricular dilation | 1 (2.0) | 4 (5.1) |
| Deep vein thrombosis | 8 (16.0) | 10 (12.8) |
| Pulmonary hypertension | 22 (44.0) | 35 (44.9) |
| Stroke | 1 (2.0) | 3 (3.8) |
| Concomitant medications, no. (%) | ||
| CYP2C9 inhibitor | 2 (4.0) | 4 (5.1) |
| CYP2C9 inducer | 0 (0.0) | 0 (0.0) |
| Statin (simvastatin) | 13 (26.0) | 15 (19.2) |
| Antibiotic | 4 (8.0) | 12 (15.4) |
| Stable warfarin dose, (mg/day) | ||
| Mean | 5.0 | 4.9 |
| Median | 4.3 | 4.3 |
| Range | 1.7–13.0 | 1.5–12.5 |
BMI, body mass index; CYP, cytochrome P450; INR, international normalized ratio.
The multivariate regression analysis model controlled for age, gender, self‐reported heritage, and concurrent statin use. We elected to include age, gender, and heritage in the model a priori, although only age and heritage were found to be significantly associated with stable warfarin dose by univariate regression analysis (Table 2). Statins were the only medication class included in the multivariate regression model, as they were associated with a 1.7 mg/day decrease in warfarin dose (P < 0.01) and > 10 customer‐owners in the cohort were using simvastatin concomitantly (Table 2). There were not enough observations of CYP2C9 inhibitors or antibiotics to include these medications in the regression model (Table 1).
Table 2.
Effects of clinical and demographic factors on stable warfarin dose by univariate linear regression analysis
| Covariate | P value | Trend in univariate linear regression |
|---|---|---|
| Heritage | P < 0.049 | −2.4 mg/day for AN |
| Age | P < 0.008 | −0.05 mg/day for increase of 1 year in age |
| Gender | P < 0.30 | +0.72 mg/day for men |
| Statin | P < 0.013 | −1.67 mg/day with concurrent statin use |
Heritage, age, gender, and statin use were all included in the multivariate regression model to assess genotype‐dose associations.
AN, Alaska Native.
The results of the analysis adjusting for age, heritage, gender, and concurrent statin use are shown in Table 3. Among the INR‐based stable warfarin dose cohort of 50 customer‐owners, VKORC1 −1639G>A and VKORC1 1173C>T were significantly associated with stable dose, decreasing the dose required to achieve therapeutic INR by 1.7 mg/day per allele (t‐test of coefficients, unadjusted P = 1.4e‐05, Bonferroni adjusted P = 7.0e‐05; Table 3). This relationship remained significant when only AN customer‐owners were included in the analysis (Bonferroni adjusted P = 3.3e‐04). VKORC1 –1639G>A and 1173C>T were in full linkage disequilibrium (Figure S1), and, thus, their effect size is the same. VKORC1 genotype explained 34% of warfarin dose variability (R 2) in the AN/AI population at SCF. The mean stable dose by VKORC1 −1639G>A reference (wildtype), heterozygote, and homozygous variant genotype was 7.5, 5.2, and 3.2 mg/day, respectively, by univariate regression analysis (P < 0.05 for each genotype comparison; Figure 2 a). CYP2C9 N218I genotype was suggestively associated with stable warfarin dose, with customer‐owners carrying one copy of the N218I allele requiring 1.1 mg/day lower warfarin dose, compared with individuals carrying two reference alleles (t‐test of coefficients, P = 0.077; Table 3). The mean stable dose for CYP2C9 N218I heterozygotes was significantly lower than reference individuals (3.7 vs. 5.1 mg/day, P = 0.005) by univariate regression analysis (Figure 2 b). Other variants identified, GGCX‐intron14, CYP4F2 g72220026G, CYP4F2*3, CYP4F11 R276C, and CYP2C9*2 and *3, were not significantly associated with warfarin dose requirement in the INR‐based cohort. CYP2C9*8, CYP2C9*11, CYP2C9*14, CYP2C9 P279T, and GGCXG421A variant alleles were not present in this cohort, and CYP2C9 M1L, CYP4F2 SpliceCG, and CYP4F2 G185V, which had been detected previously in the AN/AI population of Alaska,12 had too few observations of variant alleles in the study population to accurately assess their effect on warfarin dose. Although the combination of CYP2C9 M1L or N218I heterozygotes compared with reference individuals was not significantly associated with stable warfarin dose, the average effect trended in the expected direction by univariate analysis (−0.99 mg/day, P = 0.070) as well as by multivariate regression analysis (−0.89 mg/day, P = 0.13). No additional SNVs were found to be significantly associated with dose in a secondary analysis that adjusted for the low‐dose associated VKORC1 gene variants in the multivariate regression model. The combination of VKORC1 −1639G>A and CYP2C9 N218I, as well as age, gender, and statin use in the multivariate regression model, explained 44% of the variation in the therapeutic warfarin dose in the INR‐based dose definition cohort (adjusted R 2 = 0.38).
Table 3.
Effect of genotype on stable warfarin dose in the INR‐based cohort using multivariate regression analysis (adjusting for age, heritage, gender, and concurrent statin use) as well as univariate regression analysis for comparison
| Variant (rs ID) | Trend with stable warfarin dose (significance) | # Reference | # Heterozygotes | # Homozygote variants | Allele frequency in AN/AI at SCF (%) | |
|---|---|---|---|---|---|---|
| Multivariate analysis | Univariate analysis | |||||
| VKORC1 −1639G>A (rs9923231) | −1.7 mg/day (P = 1.4e‐05a) | −2.1 mg/day (P = 1.3e‐06a) | 9 | 25 | 16 | 59.7b |
| VKORC1 1173C>T (rs9934438) | −1.7 mg/day (P = 1.4e‐05a) | −2.1 mg/day (P = 1.3e‐06a) | 9 | 25 | 16 | 59.7b |
| CYP2C9*2 (rs1799853) | +0.43 mg/day (P = 0.58) | +0.78 mg/day (P = 0.50) | 42 | 7 | 0 | 5.2 |
| CYP2C9*3 (rs1057910) | −0.14 mg/day (P = 0.91) | +1.3 mg/day (P = 0.072) | 44 | 4 | 0 | 3.4 |
| CYP2C9 M1L (rsNA) | −0.16 mg/day (P = 0.78) | +0.16 mg/day (P = 0.66) | 49 | 1 | 0 | 1.0 |
| CYP2C9 N218I (rsNA) | −1.1 mg/day (P = 0.077) | −1.4 mg/day (P = 0.005a) | 47 | 3 | 0 | 1.4 |
| GGCX‐intron14 (rs11676382) | −0.06 mg/day (P = 0.93) | −0.68 mg/day (P = 0.43) | 44 | 6 | 0 | 3.8 |
| CYP4F2 g72220026G (rs2189784) | −0.02 mg/day (P = 0.95) | −0.02 mg/day (P = 0.95) | 22 | 21 | 7 | 31.0 |
| CYP4F2*3 (rs2108622) | −0.44 mg/day (P = 0.31) | −0.65 mg/day (P = 0.16) | 23 | 22 | 5 | 31.5 |
| CYP4F2 Splice CG (rsNA) | −3.5 mg/day (P = 3.0e‐08a) | −3.0 mg/day (P = 3.0e‐09a) | 48 | 2 | 0 | 1.4 |
| CYP4F2 G185V (rs3093153) | +1.7 mg/day (P = 0.22) | +1.4 mg/day (P = 0.37) | 47 | 2 | 1 | 2.2 |
| CYP4F11 R276C (rs8104361) | +1.1 mg/day (P = 0.22) | +1.0 mg/day (P = 0.23) | 42 | 7 | 1 | 9.1 |
AI, American Indian; AN, Alaska Native; CYP, cytochrome P450; GGCX, gamma‐glutamyl carboxylase; INR, international normalized ratio; SCF, Southcentral Foundation; VKORC1, vitamin K oxidoreductase complex 1.
The P value remains significant after adjusting for multiple testing using Bonferroni correction. Variant allele frequency from an AN/AI population at SCF is included for comparison (Fohner et al.12). CYP2C9*8, CYP2C9*11, CYP2C9*14, CYP2C9 P279T, or GGCXG421A variant alleles were not present in the INR‐based dose cohort. The reported allele frequency is for the variant allele, an alternative allele to the reference allele, which is defined by the global population.
The variant allele frequency in the AN/AI population is the major allele for the VKORC1 −1639G>A and 1173C>T.
Figure 2.
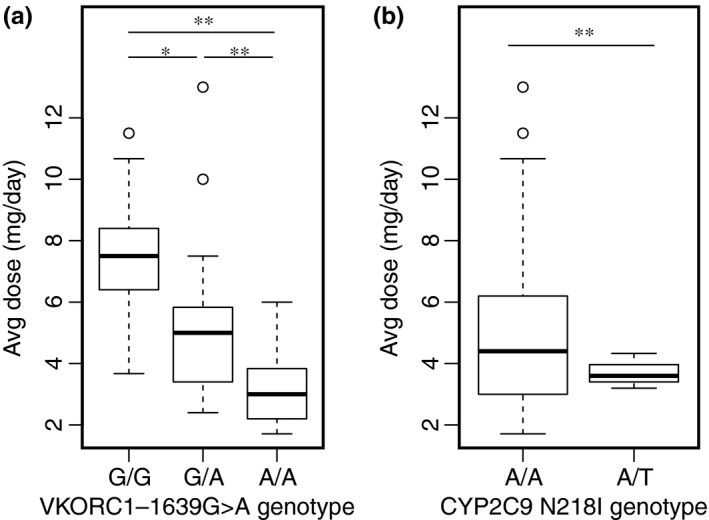
Effect of (a) vitamin K epoxide oxidase reductase complex subunit 1 (VKORC1) and (b) cytochrome P450 (CYP)2C9 N218I genotype on warfarin dose, assessed by univariate regression analysis in the international normalized ratio‐based cohort. *Denotes P < 0.05 and **P < 0.01 by t‐test of coefficients.
Interestingly, AN customer‐owners required a significantly lower daily warfarin dose to achieve therapeutic INR (4.6 mg/day), compared with those of AI heritage (7.0 mg/day; t‐test of coefficients, P = 0.049; Figure 3). However, this was well explained by a substantial difference in the diagnostic VKORC1 allele frequencies in these heritage groups, with heritage losing statistical significance in the multivariate regression model when evaluating the gene‐dose effect from VKORC1. The minor allele frequency (MAF) of the low‐dose associated VKORC1 −1639G>A variant was 61.6% in the AN population, compared with 28.6% in AI customer‐owners in this study cohort.
Figure 3.
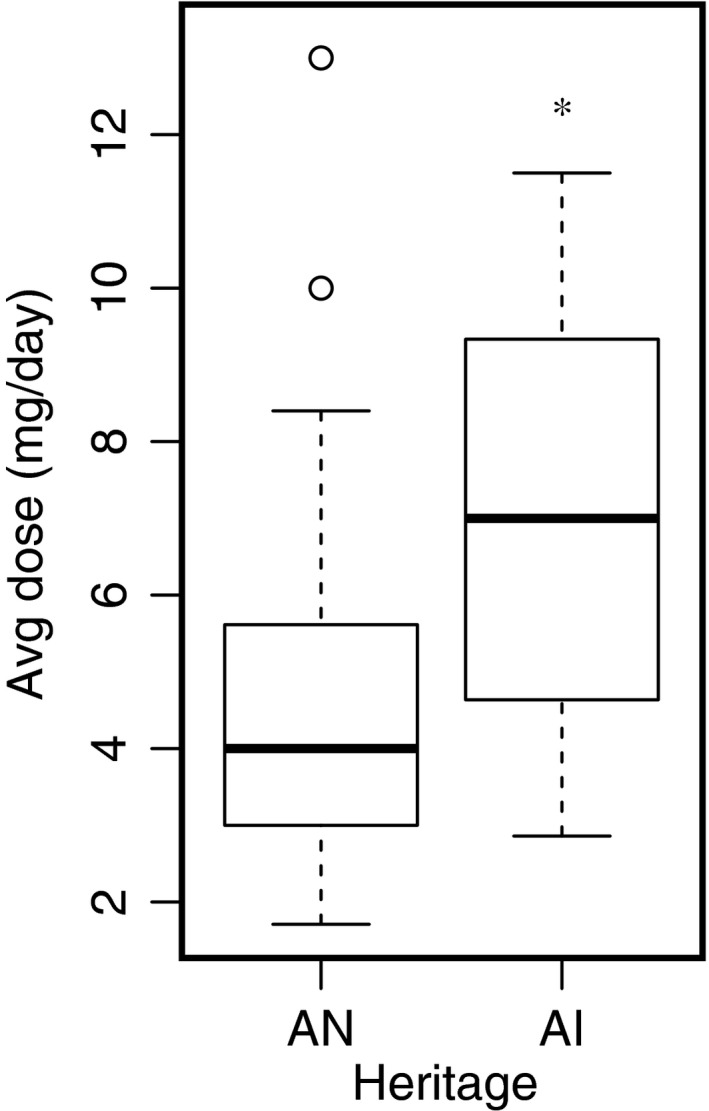
Effect of self‐reported heritage on warfarin dose, assessed by univariate regression analysis in the international normalized ratio‐based cohort. The Alaska Native (AN) subgroup includes the following tribes: Inupiaq, Athabascan subgroups, Tlingit, Tsimshian, Haida, Eyak, Aleut/Unangan, Central Yup'ik, Cup'ik, and Sugpiaq/Alutiiq. The American Indian (AI) subgroup includes participants affiliated with tribes in the lower 48 states. *Denotes P < 0.05 by t‐test of coefficients, compared with the AN subgroup.
Analysis of the consecutive (non‐INR‐based) warfarin dose definition in 78 customer‐owners showed similar statistical trends. VKORC1 −1639G>A and VKORC1 1173C>T remained significantly associated with a lower warfarin dose requirement (P < 0.05), as was CYP2C9 N218I (P < 0.05; Table 4). Although not found to be associated with warfarin dose in the INR‐based cohort, GGCX‐intron14 was suggestively associated (P = 0.087) with lower warfarin dose, compared with reference in the consecutive dose definition cohort. Again, CYP2C9*8, CYP2C9*14, and CYP2C9 P279T variant alleles were not present, whereas CYP2C9*11, CYP2C9 M1L, GGCX G421A, and CYP4F2 SpliceCG had too few observations of variant alleles to accurately assess their effect on dose. CYP4F2 g72220026G, CYP4F2*3, CYP4F2 G185V, CYP4F11 R276C, and CYP2C9*2 and *3 did not seem to be associated with warfarin dose requirement. No additional SNVs were found to be significantly associated with dose in a secondary analysis that adjusted for the low‐dose associated VKORC1 gene variants in the multivariate regression model.
Table 4.
Effect of genotype on stable warfarin dose in the consecutive (non‐INR‐based) cohort using multivariate regression analysis (adjusting for age, heritage, gender, and concurrent statin use) as well as univariate regression analysis for comparison
| Variant (rs ID) | Trend with stable warfarin dose (significance) | # Reference | # Heterozygotes | # Homozygote Variants | Allele frequency in AN/AI at SCF (%) | |
|---|---|---|---|---|---|---|
| Multivariate analysis | Univariate analysis | |||||
| VKORC1 −1639G>A (rs9923231) | −1.8 mg/day (P = 1.6e‐08a) | −1.8 mg/day (P = 8.1e‐08a) | 14 | 43 | 21 | 59.7b |
| VKORC1 1173C>T (rs9934438) | −1.8 mg/day (P = 1.6e‐08a) | −1.8 mg/day (P = 8.1e‐08a) | 14 | 43 | 21 | 59.7b |
| CYP2C9*2 (rs1799853) | +0.16 mg/day (P = 0.80) | +0.15 mg/day (P = 0.83) | 68 | 9 | 0 | 5.2 |
| CYP2C9*3 (rs1057910) | −0.46 mg/day (P = 0.61) | +0.03 mg/day (P = 0.97) | 69 | 7 | 0 | 3.4 |
| CYP2C9*11 (rs28371685) | −1.0 mg/day (P = 0.29) | +0.07 mg/day (P = 0.79) | 77 | 1 | 0 | 0.0 |
| CYP2C9 M1L (rsNA) | +0.37 mg/day (P = 0.47) | +0.50 mg/day (P = 0.06) | 77 | 1 | 0 | 1.0 |
| CYP2C9 N218I (rsNA) | −1.1 mg/day (P = 0.014) | −1.3 mg/day (P = 0.001a) | 75 | 3 | 0 | 1.4 |
| GGCX‐intron14 (rs11676382) | −1.2 mg/day (P = 0.087) | −1.3 mg/day (P = 0.039) | 69 | 9 | 0 | 3.8 |
| GGCX G421A (rsNA) | +0.25 mg/day (P = 0.61) | +0.05 mg/day (P = 0.84) | 76 | 1 | 0 | 0.6 |
| CYP4F2 g72220026G (rs2189784) | +0.27 mg/day (P = 0.40) | +0.22 mg/day (P = 0.53) | 35 | 32 | 11 | 31.0 |
| CYP4F2*3 (rs2108622) | −0.25 mg/day (P = 0.48) | −0.35 mg/day (P = 0.34) | 35 | 36 | 6 | 31.5 |
| CYP4F2 Splice CG (rsNA) | −2.3 mg/day (P = 1.2e‐04a) | −2.2 mg/day (P = 3.7e‐06a) | 74 | 3 | 0 | 1.4 |
| CYP4F2 G185V (rs3093153) | +0.72 mg/day (P = 0.49) | +0.63 mg/day (P = 0.57) | 73 | 4 | 1 | 2.2 |
| CYP4F11 R276C (rs8104361) | +0.57 mg/day (P = 0.35) | +0.58 mg/day (P = 0.34) | 63 | 14 | 1 | 9.1 |
AI, American Indian; AN, Alaska Native; CYP, cytochrome P450; GGCX, gamma‐glutamyl carboxylase; INR, international normalized ratio; SCF, Southcentral Foundation; VKORC1, vitamin K oxidoreductase complex 1.
The P value remains significant after adjusting for multiple testing using Bonferroni correction. Variant allele frequency from an AN/AI population at SCF is included for comparison (Fohner et al.12). CYP2C9*8, CYP2C9*14, or CYP2C9 P279T variant alleles were not present in the consecutive (non‐INR‐based) dose cohort. The reported allele frequency is for the variant allele, an alternative allele to the reference allele, which is defined by the global population.
The variant allele frequency in the AN/AI population is the major allele for the VKORC1 −1639G>A and 1173C>T.
Discussion
Warfarin, widely used to prevent thromboembolic events and stroke, has been the focus of many pharmacogenetic studies; however, there are few data specifically addressing the clinical implications of genetic variation in the AN/AI populations.10, 11 To our knowledge, this is the first study to evaluate genotype–phenotype associations for warfarin pharmacogenes with stable warfarin dose in an AN/AI population. Results of this retrospective study suggest that: (i) VKORC1 genotype is strongly associated with stable warfarin dose in the AN/AI population, explaining 34% of the variability in a therapeutic warfarin dose (R 2), somewhat higher than that seen in other populations worldwide; (ii) AN people require a lower warfarin dose than AI people on average, explained by a difference in the diagnostic VKORC1 allele frequencies; (iii) the novel CYP2C9 variant, N218I, lowers warfarin dose requirement to a clinically meaningful degree. The combination of VKORC1 −1639G>A and CYP2C9 N218I, as well as age, gender, and statin use, explained 44% of the variation in the therapeutic warfarin dose, establishing the clinical validity of pharmacogenetic associations in this population. Other factors for consideration of clinical utility at SCF include cost, access to genetic testing, and impact of the current empirical approach to warfarin dosing when many of the customer‐owners live in remote communities throughout the state.
The significantly lower warfarin dose in AN compared with AI customer‐owners was well explained by a higher frequency of the low‐dose associated VKORC1 alleles (−1639G>A and 1173C>T) in the AN heritage group. Although heritage was associated with stable warfarin dose in a univariate regression model, it was no longer a statistically significant adjustment in the multivariate regression model when evaluating the additive gene‐dose effect from VKORC1. BMI between the two heritage groups did not greatly differ, with the mean BMI for the AN and AI customer‐owners being 30.7 and 30.5, respectively. Racial/ethnic differences in drug response can arise from multiple nongenetic factors (e.g., diet and environment), but in the case of warfarin it would seem that VKORC1 variation is the predominant determinant.
The percent of warfarin dose variability explained by VKORC1 genotype has previously been shown to differ by race, due to differences in VKORC1 MAFs across racial groups. The low‐dose VKORC1 alleles were found in the highest frequency in East Asian populations and associated with a lower average dose requirement than populations of European or African origin.7, 16 The VKORC1 alleles were also significantly associated with a decrease in dose for both European Americans and African Americans, but with a greater effect in Europeans who had a higher frequency of the VKORC1 variant allele, compared with African Americans.17
The relatively small sample size likely reduced our ability to find significant associations with the well‐established CYP2C9 variants (i.e., *2 and *3), although we did find that the CYP2C9*3 allele was associated with a lower warfarin dose requirement: 0.14 mg/day for INR‐based stable dose and 0.46 mg/day for the consecutive (non‐INR‐based) stable dose definition. In the absence of more definitive data, we hypothesize that those alleles would have the expected effect (reduced dose requirement) on average in the SCF population. Although CYP2C9*2 and *3 are predictive of a lower warfarin dose in other world populations, they have low MAFs in the AN/AI populations of Alaska (5.2 and 3.4%, respectively, Fohner et al.12), and so their contribution to stable warfarin dose in this population is likely to be minor.
Novel variants unique to the AN/AI populations, such as CYP2C9 M1L or N218I, may be more important to consider. Although only three AN customer‐owners were heterozygous for CYP2C9 N218I, the change is predicted to be disruptive to CYP2C9 enzyme function, as it resulted in a lower warfarin dose in both the INR‐based and consecutive stable warfarin dose cohorts. Unpublished findings from our research team support a causal association, with the recombinant N218I variant enzyme purified from Escherichia coli exhibiting ~ 70% reduction in intrinsic clearance to the major 7‐hydroxy and 6‐hydroxy warfarin metabolites, compared with wildtype CYP2C9 protein (McDonald et al., unpublished results, 2018).
The CYP4F2*3 allele has been associated with reduced hepatic concentrations of the CYP4F2 enzyme, decreasing vitamin K catabolism,18 and was, therefore, expected to confer an increase in stable warfarin dose requirement. This was not the case. Possible reasons for deviation from the expected result include uncontrolled dietary effects (e.g., consumption of vitamin K–rich foods) or possibly a difference in the penetrance of the CYP4F2*3 allele because of other uncontrolled genetic and regulatory factors.19
With regard to the significant covariates, statins have been shown to decrease the stable warfarin dose requirement, although it is unclear how they may be involved in the causal pathway of warfarin response. Possible explanations are that they inhibit organic anion transporter 2 mediated uptake of warfarin into hepatocytes20 or that they affect the composition of lipoproteins that facilitate the trafficking of vitamin K into and out of hepatocytes.21 As expected, increasing age was significantly associated with a reduction in warfarin dose requirement, reflecting a variety of anatomic and biochemical changes that occur with aging and that can affect drug disposition and response.22 The lack of a significant association with gender may be attributed to its small effect size and the relatively small sample size of the study, as a causal relationship is expected based on the bulk of the published literature.
The main limitation of this study is the relatively small number of customer‐owners included in the primary analysis; however, a small sample size is inherent to the study population. There are ~ 113,000 AN/AI people living in Alaska,23 with only a fraction of those receiving warfarin anticoagulation therapy. Nonetheless, it is important to conduct studies such as ours, with its inherent limitations, to enhance the knowledge base, guide future research, and support the introduction of precision medicine, where appropriate, for this historically underserved population. The primary INR‐based stable dose definition was selected to allow for a more rigorous dose tied to therapeutic INR, increasing precision, but it also reduced the sample size appreciably, decreasing the study's power. By excluding customer‐owners who were not receiving warfarin chronically (at least 6 months), we likely excluded individuals being treated for deep venous thrombosis and pulmonary embolism, as these indications typically require 3 months of warfarin therapy, whereas atrial fibrillation or mechanical valves require life‐long prophylaxis treatment. This allowed for the analysis of customer‐owners receiving chronic warfarin therapy instead of the initial dosing period, in which the warfarin dose is often unstable,24 and during which individuals carrying CYP2C9*2 or *3 variant alleles require additional dose adjustments and take longer to achieve a stable, therapeutic warfarin dose.5 However, a consequence of the small study sample size is also that there were fewer AI customer‐owners included in the study, so most of the observed variation is due to the AN population. More AI customer‐owners are needed to inform on MAFs specific to the AI population and their effect on warfarin dose. To address this limitation, we included the consecutive (non‐INR‐based) stable dose definition in an effort to include more customer‐owners, and AI individuals, in the final study analysis. The observation of a higher mean therapeutic warfarin dose in customer‐owners of AI heritage, compared with AN, in both the INR‐based and consecutive dose cohorts is consistent with years of clinical experience by the SCF anticoagulation team. Future work should aim to analyze the effect of genetic variation across the different tribes in the lower 48 states, rather than grouping them together under AI heritage.
In conclusion, in an AN/AI population, VKORC1 –1639G>A was significantly associated with a lower stable warfarin dose, and a novel CYP2C9 gene variant (N218I) was suggestively associated with a clinically relevant lower dose. These findings could potentially be incorporated into pharmacogenetic screening to help guide dose selection to improve warfarin safety and efficacy for this rural population. These observations and marked differences in allele frequencies found in AN/AI subgroups12 compels further investigation in order to better understand the role of genetic variation in warfarin therapy outcomes and potentially optimize personalized medicine for this underserved population. The AN/AI population must be adequately represented in pharmacogenetic studies that assess genotype–phenotype associations in order to potentially inform on the dosing of medications with established clinical associations with pharmacogene variation, especially narrow therapeutic index substrates, such as warfarin.
Funding
National Institutes of Health (U01 GM092676, P01 GM116691, P30 ES007033, T32 GM007750).
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
L.M.H., R.F.R., K.E.T., T.A.T., and D.L.V. wrote the manuscript. L.M.H., R.F.R., D.A.D., B.D.S., A.E.R., K.E.T., T.A.T., and D.L.V. designed the research. L.M.H., R.F.R., B.D.S., M.M., P.L.J., and A.E.F. performed the research. L.M.H., L.R., T.L., and T.A.T. analyzed the data.
Supporting information
Figure S1. Linkage disequilibrium (LD) in the VKORC1 locus for rs9923231 (−1639G>A) and rs9934438 (1173C>T) SNVs in all 118 genotyped customer‐owners from SCF. R 2 was 1.0 for this variant pair.
Acknowledgments
The authors would like to thank Barbara Kavanaugh, Program Manager for the NWA‐PGRN, for directing programmatic collaborations, LT Christopher Chong, PharmD, and Austen Rodgers, who collected blood samples at SCF, primary care staff who assisted with study coordination and, most of all, the customer‐owners of SCF who participated in the study.
[Correction added on May 10, 2019, after first online publication: Burhan A. Khan was added as fourth author.]
[The copyright line for this article was changed on March 18, 2019 after original online publication.]
References
- 1. Coumadin (warfarin sodium) [package insert]. (Bristol‐Myers Squibb, Princeton, NJ, 2017). [Google Scholar]
- 2. Zirlik, A. & Bode, C. Vitamin K antagonists: relative strengths and weaknesses vs. direct oral anticoagulants for stroke prevention in patients with atrial fibrillation. J. Thromb. Thrombolysis 43, 365–379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rettie, A.E. & Tai, G. The pharmacogenomics of warfarin: closing in on personalized medicine. Mol. Interv. 6, 223–227 (2006). [DOI] [PubMed] [Google Scholar]
- 4. Gage, B.F. , Eby, C. , Milligan, P.E. , Banet, G.A. , Duncan, J.R. & McLeod, H.L. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb. Haemost. 91, 87–94 (2004). [DOI] [PubMed] [Google Scholar]
- 5. Higashi, M.K. et al Association between CYP2C9 genetic variants and anticoagulation‐related outcomes during warfarin therapy. JAMA 287, 1690–1698 (2002). [DOI] [PubMed] [Google Scholar]
- 6. Limdi, N.A. et al Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African‐American and European‐American patients on warfarin. Clin. Pharmacol. Ther. 83, 312–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rieder, M.J. et al Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N. Engl. J. Med. 352, 2285–2293 (2005). [DOI] [PubMed] [Google Scholar]
- 8. Caldwell, M.D. et al CYP4F2 genetic variant alters required warfarin dose. Blood 111, 4106–4112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rieder, M.J. , Reiner, A.P. & Rettie, A.E. Gamma‐glutamyl carboxylase (GGCX) tagSNPs have limited utility for predicting warfarin maintenance dose. J. Thromb. Haemost. 5, 2227–2234 (2007). [DOI] [PubMed] [Google Scholar]
- 10. Henderson, L.M. et al P450 pharmacogenetics in indigenous North American populations. J. Pers. Med. 8, E9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaja, C. , Burke, W. , Thummel, K. , Edwards, K. & Veenstra, D.L. Cytochrome p450 enzyme polymorphism frequency in indigenous and Native American populations: a systematic review. Community Genet. 11, 141–149 (2008). [DOI] [PubMed] [Google Scholar]
- 12. Fohner, A.E. et al Variation in genes controlling warfarin disposition and response in American Indian and Alaska Native people: CYP2C9, VKORC1, CYP4F2, CYP4F11, GGCX. Pharmacogenet. Genomics 25, 343–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edson, K.Z. et al Cytochrome P450‐dependent catabolism of vitamin K: ω‐hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry 52, 8276–8285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrett, J.C. , Fry, B. , Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- 15. Klein, T.E. et al Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 360, 753–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abecasis, G.R. et al An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Limdi, N.A. et al Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 115, 3827–3834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McDonald, M.G. , Rieder, M.J. , Nakano, M. , Hsia, C.K. & Rettie, A.E. CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol. Pharmacol. 75, 1337–1346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shendre, A. et al Race‐specific influence of CYP4F2 on dose and risk of hemorrhage among warfarin users. Pharmacotherapy 36, 263–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bi, Y.A. et al Role of hepatic organic anion transporter 2 in the pharmacokinetics of R‐ and S‐warfarin. In vitro studies and mechanistic evaluation. Mol. Pharm. 15, 1284–1295 (2018). [DOI] [PubMed] [Google Scholar]
- 21. Shearer, M.J. & Newman, P. Metabolism and cell biology of vitamin K. Thromb. Haemost. 100, 530–547 (2008). [PubMed] [Google Scholar]
- 22. Shendre, A. , Parmar, G.M. , Dillon, C. , Beasley, T.M. & Limdi, N.A. Influence of age on warfarin dose, anticoagulation control, and risk of hemorrhage. Pharmacotherapy 38, 588–596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norris, T. , Vines, P.L. & Hoeffel, E.M . The American Indian and Alaska Native Population: 2010 In: 2010 Census Briefs (US Department of Commerce, Economics and Statistics Administration, US Census Bureau, Suitland, MD, 2012). [Google Scholar]
- 24. Beyth, R.J. , Milligan, P.E. & Gage, B.F. Risk factors for bleeding in patients taking coumarins. Curr. Hematol. Rep. 1, 41–49 (2002). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Linkage disequilibrium (LD) in the VKORC1 locus for rs9923231 (−1639G>A) and rs9934438 (1173C>T) SNVs in all 118 genotyped customer‐owners from SCF. R 2 was 1.0 for this variant pair.


