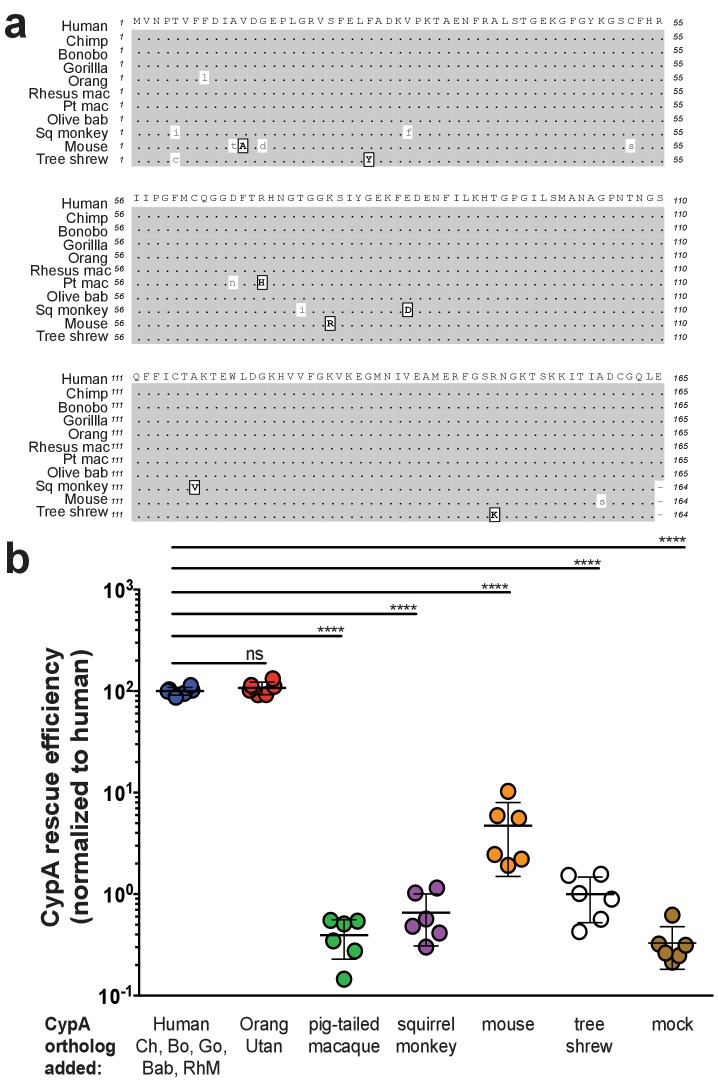
Figure 1. Murine CypA has a diminished ability to facilitate HCV replication.
(A) An amino acid sequence alignment of CypA from diverse species. Similar amino acids are indicated in boxed, bold capital letters while differences are lowercase. Species are arranged from top to bottom in increasing evolutionary distance from human. For pigtailed macaque, all experiments utilized a TRIM5-CypA fusion – only the residues of the CypA portion of the fusion are depicted here. (B) Huh7.5 cells expressing an shRNA against endogenous human CypA (Huh7.5-shRNA CypA) were transduced to express different CypA orthologs and then infected with a HCV reporter genome expressing secreted Gaussia luciferase (Jc1-Gluc, MOI = 0.1). At five dpi, the luciferase activity of the supernatants was assessed as a proxy for viral replication. CypA rescue efficiency is shown normalized to Huh7.5-shRNA CypA transduced with human CypA, which is 100% identical at the amino acid level to chimpanzee, bonobo, gorilla, olive baboon and rhesus macaque CypA. Results shown are from two representative experiments, each with triplicate samples. Lines and error bars represent the mean ± SD. Ordinary two-way ANOVA test performed followed by Dunnett’s multiple comparisons test with all means compared to that of the +human CypA line. Chimp/Ch, chimpanzee; Bo, bonobo; Go, gorilla; Orang, orangutan; Rhesus mac/RhM, rhesus macaque; Pt mac, pigtailed macaque; Olive bab/Bab, olive baboon; Sq monkey, squirrel monkey. ****, p<0.0001; ns, not significant.