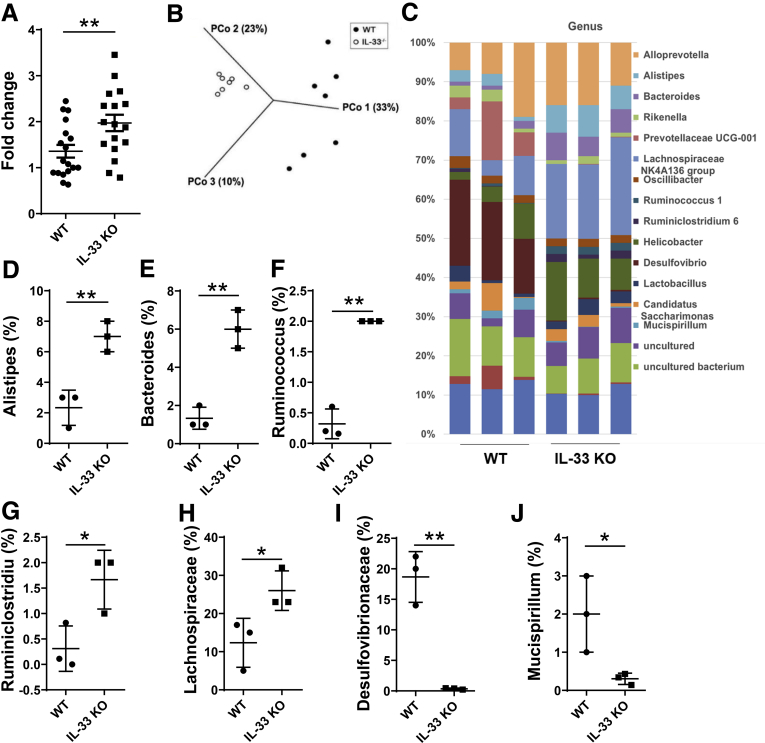
Figure 6.
Increased total gut microbiota and altered composition of gut bacterial communities in IL33-/-mice. Fecal bacterial DNA was prepared from WT and IL33-/- mice. (A) Total bacteria (n = 17 mice/group) were determined by measuring 16S rDNA using qRT-PCR, and data was normalized to eukaryotic β-actin. (B, C) 16S rRNA pyrosequencing was performed. Principal Coordinates Analysis of unweighted UniFrac distances for 16S rRNA V3–V4 sequence data of gut microbiota in WT and IL33-/- mice (n = 7/group) (B). Differences in the microbial distributions of WT and IL33-/- mice (n = 3/group) were found at the genus level (C). The percentages of Alistipes (D), Bacteroides (E), Ruminococcus (F), Ruminiclostridium (G), Lachnospiraceae NK4A136 group (H), Desulfovibrio (I), and Mucispirillum (J) in the total bacteria in the feces of WT and IL33-/- mice, respectively. Significance was calculated using Student t test. *P < .05. **P < .01. Data are represented as means ± standard deviation.