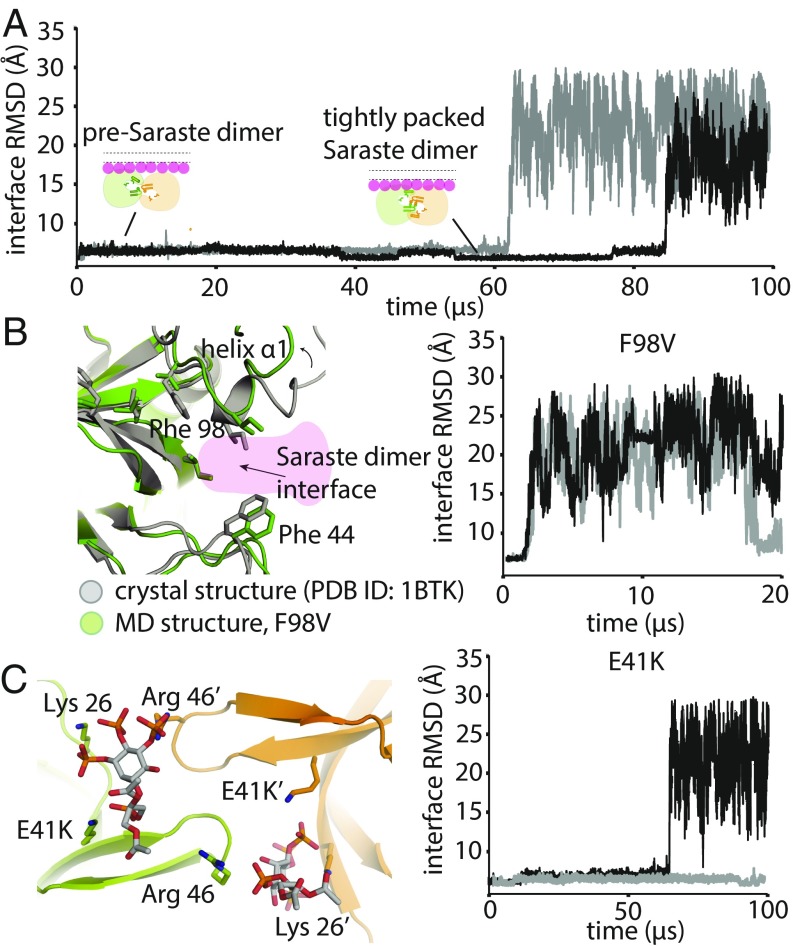
Fig. 4.
Stability analysis of Saraste dimer variants on a membrane. (A) Representative tempered binding simulation trajectories showing dissociation of the tightly packed Saraste conformation in two individual trajectories. The tempered binding simulations started from a Saraste dimer on a membrane containing 6% PIP3 and 94% POPC. (B) Stability analysis of the F98V mutant, starting from the Saraste dimer conformation, on a membrane. An instantaneous structure from a simulation (t = 20 μs) shows the local conformational rearrangement that occurred at the Saraste interface before the dimer dissociated. Representative tempered binding simulation trajectories show the dissociation of the F98V dimer in two individual trajectories. (C) Stability analysis of the E41K mutant, starting from the Saraste dimer conformation, on a membrane. An instantaneous structure from a simulation (t = 100 μs) shows PIP3 bound to residue Lys-41 at the bridging site before the dimer dissociated. Representative tempered binding simulation trajectories show the dissociation of the E41K dimer in two individual trajectories.