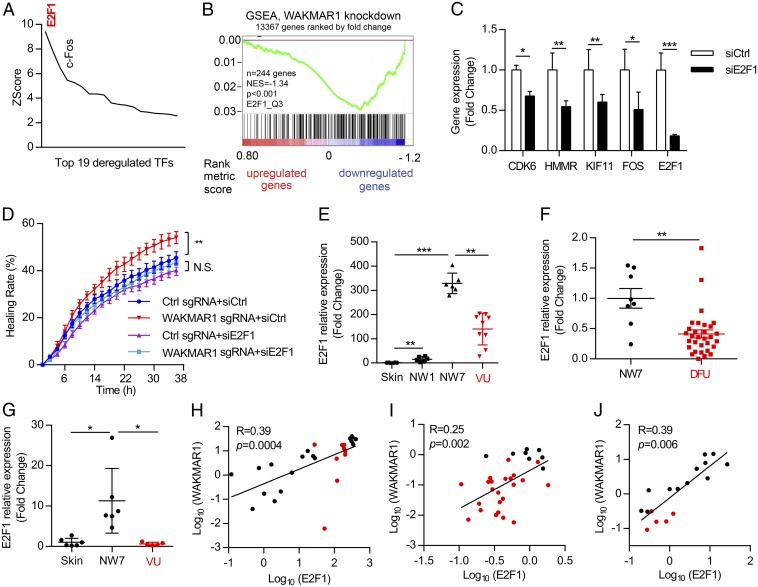
Fig. 5.
E2F1 acts as an upstream regulator in the WAKMAR1-regulated gene network. (A) MetaCore analysis identified transcription factors (TFs) with overrepresented binding sites among WAKMAR1-regulated genes. Only TFs with changed expression after WAKMAR1 knockdown are shown. (B) GSEA evaluated the enrichment of E2F1-target genes in the WAKMAR1-regulated genes. NES, normalized enrichment score. (C) qPCR analysis of CDK6, HMMR, KIF11, FOS, and E2F1 in keratinocytes transfected with E2F1 siRNA for 24 h (n = 3). (D) Scratch wound assay of keratinocytes after WAKMAR1 knockdown and/or E2F1 silencing (n = 8). qPCR analyses of E2F1 in the skin in day 1 (NW1) and day 7 (NW7) normal wounds from six healthy donors and in the wound edges of nine patients with VU (E and H) in NW7 (n = 8) vs. DFU (n = 29) (F and I) and in wound-edge epidermis isolated from healthy skin and NW7 (n = 6) and VU (n = 5) with LCM (G and J) are shown. (H–J) Expression correlation of WAKMAR1 with E2F1 in human skin, normal wounds (black dots), and chronic wounds (red dots). *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired two-tailed Student’s t test (C), two way-ANOVA (D), Mann–Whitney U test (E–G), or Pearson’s correlation test (H–J). N.S., not significant. Data are presented as mean ± SD (C) or mean ± SEM (D–G).