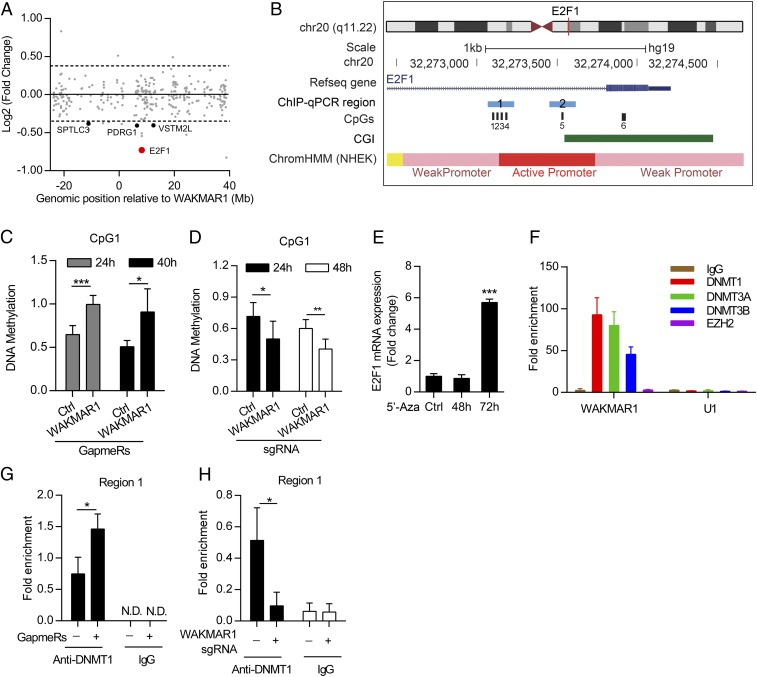
Fig. 6.
WAKMAR1 activates E2F1 expression by suppressing E2F1 promoter methylation. (A) Chr20 gene expression in keratinocytes with WAKMAR1 knockdown. The gene start distance from the WAKMAR1 transcriptional start site is shown on the x axis. The log-twofold change of expression levels between control and WAKMAR1 knockdown is shown on the y axis. Genes with absolute fold change ≥ 1.3 and P < 0.05 are highlighted. (B) Genomic snapshot of E2F1 promoter: CpG sites (black bars) analyzed by MSRE-qPCR (CpG1, CpG5, and CpG6) and bisulfite pyrosequencing (CpG1, CpG2, CpG3, and CpG4) and regions analyzed by ChIP-qPCR (blue bars) are highlighted. CGI, CpG island; ChromHMM, chromatin state segmentation by hidden Markov model from ENCODE/Broad; NHEK, normal human epidermal keratinocytes. MSRE-qPCR analyses of DNA methylation at CpG1 in keratinocytes transfected with WAKMAR1 GapmeR for 24 h and 40 h (n = 6) (C) or CRISPR/Cas9-SAM plasmids for 24 h and 48 h (n = 6) (D) are shown. Ctrl, control. (E) qPCR of E2F1 in keratinocytes treated with 5′-Aza-2′-deoxycytidine (5′-Aza) (n = 3). (F) qPCR analysis of WAKMAR1 and U1 small nuclear RNA immunoprecipitated from keratinocytes with DNMT1, DNMT3A, DNMT3B, and EZH2 antibodies or IgG (n = 3). ChIP-qPCR of E2F1 promoter region 1 was performed in keratinocytes transfected with WAKMAR1 GapmeRs (G) or CRISPR/Cas9-SAM plasmids (H) and immunoprecipitated using DNMT1 antibody or IgG (n = 3). *P < 0.05; ***P < 0.001 by unpaired two-tailed Student’s t test (C–E, G, and H). N.D., not detected. Data are presented as mean ± SD.