Significance
Rice seeds comprised of embryo and endosperm are generated when the vegetative cell pollen tube delivers two sperm cells that fertilize central and egg cells, respectively. We show that the ROS1a DNA glycosylase actively demethylates DNA of rice vegetative cell genomes, which is required for viable seed production. Thus, DNA glycosylase-mediated DNA demethylation, conserved for 150 million years between rice and Arabidopsis, is likely an essential feature of seed development in all flowering plants. We also reveal that global DNA methylation levels in rice sperm and egg cells are different both from each other and also compared with the embryo, suggesting that dynamic DNA methylation reprogramming occurs during plant embryogenesis.
Keywords: DNA methylation, DNA demethylation, epigenetics, rice, pollen
Abstract
Epigenetic reprogramming is required for proper regulation of gene expression in eukaryotic organisms. In Arabidopsis, active DNA demethylation is crucial for seed viability, pollen function, and successful reproduction. The DEMETER (DME) DNA glycosylase initiates localized DNA demethylation in vegetative and central cells, so-called companion cells that are adjacent to sperm and egg gametes, respectively. In rice, the central cell genome displays local DNA hypomethylation, suggesting that active DNA demethylation also occurs in rice; however, the enzyme responsible for this process is unknown. One candidate is the rice REPRESSOR OF SILENCING 1a (ROS1a) gene, which is related to DME and is essential for rice seed viability and pollen function. Here, we report genome-wide analyses of DNA methylation in wild-type and ros1a mutant sperm and vegetative cells. We find that the rice vegetative cell genome is locally hypomethylated compared with sperm by a process that requires ROS1a activity. We show that many ROS1a target sequences in the vegetative cell are hypomethylated in the rice central cell, suggesting that ROS1a also demethylates the central cell genome. Similar to Arabidopsis, we show that sperm non-CG methylation is indirectly promoted by DNA demethylation in the vegetative cell. These results reveal that DNA glycosylase-mediated DNA demethylation processes are conserved in Arabidopsis and rice, plant species that diverged 150 million years ago. Finally, although global non-CG methylation levels of sperm and egg differ, the maternal and paternal embryo genomes show similar non-CG methylation levels, suggesting that rice gamete genomes undergo dynamic DNA methylation reprogramming after cell fusion.
Plant haploid gametes, sperm and egg, are generated by meiosis in male and female gametophytes, respectively. Vegetative and central cells, adjacent to the sperm and egg cells, respectively, are necessary for fertilization and seed development. The vegetative cell in pollen generates a pollen tube that transports two sperm cells to the ovary. The egg is fertilized by one sperm to form the embryo, and the homodiploid central cell is fertilized by the other sperm cell to generate the triploid endosperm, a nutrient-rich tissue that feeds the growing embryo or the seedling. Monocot cereal seeds provide 50% of the world’s dietary energy consumption, and most calories are in the endosperm (1). Rice feeds half of the global population and is the predominant source of nutrition for the world’s poor (2). Understanding proper development of rice companion cells, gametes, and seeds is key to improvement of crop security worldwide.
DNA methylation is associated with transcription silencing in eukaryotic organisms (3). In plants, methylation is in three nucleotide contexts: CG, CHG, and CHH (H = A, T, or C) (4). In Arabidopsis, CG methylation is maintained by DNA METHYLTRANSFERASE 1 (MET1) and is present in both genes and transposable elements (TEs) (4). Non-CG methylation is present in TEs and is maintained by distinct pathways depending on the chromatin environment (heterochromatic vs. euchromatic) (4). CHG and CHH methylation in short euchromatic TEs with a low level of heterochromatic histone marks (e.g., H3K9me2) is maintained by the RNA-dependent DNA methylation (RdDM) pathway, which uses mobile small RNAs (sRNAs) to recruit the DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) DNA methyltransferase to target sequences (4, 5). Non-CG methylation in heterochromatic TEs with higher levels of heterochromatic marks is maintained by CHROMOMETHYLASE (CMT) 3 (CHG) and CMT2 (CHH) DNA methyltransferases (4, 5). DNA methylation is actively removed by a family of DNA glycosylases, DME, ROS1, and DME-LIKE (DML) proteins 2 and 3 in Arabidopsis (6), which excise 5-methylcytosine that is replaced by cytosine via the base excision repair pathway.
DME-mediated DNA demethylation is essential for Arabidopsis plant reproduction, and inheritance of loss-of-function maternal or paternal mutant dme alleles results in seed abortion or reduced sperm transmission, respectively (7, 8). DME is expressed in the vegetative and central cells and demethylates their genomes at about 10,000 sites, primarily at euchromatic TEs and the edges of large TEs (3, 9–12). DNA demethylation at central cell TEs regulates adjacent gene expression, which can result in gene imprinting in the endosperm (13). By contrast, ROS1 and DML-mediated DNA demethylation are not essential for Arabidopsis reproduction (14). ROS1 and DML genes are expressed primarily in sporophytic (e.g., roots and shoots) cells and at a lower level compared with DME in the vegetative cell (15, 16).
Phylogenetic analysis identified rice DNA demethylation genes only in the ROS1 and DML orthology group (17). Rice ROS1a, like DME in Arabidopsis, is expressed in pollen and unfertilized ovules, and its loss-of-function mutation results in seed abortion and reduced sperm transmission (18). However, whether ROS1a has DNA demethylation function during rice reproduction is unknown.
Here, we report the role of DNA demethylation by ROS1a in rice male gametophytes. We found that rice wild-type vegetative cells are CG-hypomethylated at specific loci compared with sperm. The hypomethylation is lost in ros1a mutant vegetative cells, indicating that ROS1a is responsible for DNA demethylation in the vegetative cell. ROS1a targets in the vegetative cell were also hypomethylated in the central cell and maternal endosperm genomes, suggesting that ROS1a may function in the central cell. ROS1a is required for non-CG hypermethylation in sperm at hypomethylated sites in the vegetative cell, which may involve communication between the vegetative and sperm cells to reinforce methylation at sperm TEs. Last, we observed that sperm and egg non-CG methylation is dynamically reprogrammed during embryogenesis. Our findings reveal that DNA glycosylase-mediated active DNA demethylation in male gametogenesis is catalyzed by ROS1a and that this mechanism has been conserved in monocots and dicots, despite 150 million years of divergent evolution (19).
Results
Local Hypomethylation Occurs in Rice Vegetative Cells.
To compare the DNA methylation patterns of sperm and vegetative cells in rice, we manually isolated sperm cells and vegetative cell nuclei from Nipponbare. The plants we used ubiquitously express an H2B-GFP transgene (20) that facilitated purification of vegetative cell nuclei visualized under fluorescence microscopy (SI Appendix, Fig. S1). Three hundred forty-one purified sperm cells and 432 vegetative cell nuclei were used to generate genome-wide maps (11-fold and ninefold genome coverage, respectively) of DNA methylation (SI Appendix, Table S1), as previously described (9, 21).
We observed similar global shapes of sperm and vegetative cell DNA methylation at TEs and genes with varying overall DNA methylation levels (SI Appendix, Fig. S2). Sperm and vegetative cells were both highly CG-methylated in genes and TEs with sperm methylation levels being slightly higher than the vegetative cell levels (SI Appendix, Fig. S2 A and D). For non-CG methylation, the vegetative cell showed higher DNA methylation than the sperm in TEs (SI Appendix, Fig. S2 B and C). This observation is also found in Arabidopsis, suggesting the highly dimorphic non-CG methylation levels and chromatin dynamics are conserved during late male gametophyte development when a single cell division generates the vegetative cell and sperm lineage (10). In genes, the non-CG methylation levels were very low (SI Appendix, Fig. S2 E and F), similar to other cell types and tissues previously reported in plants (5, 17).
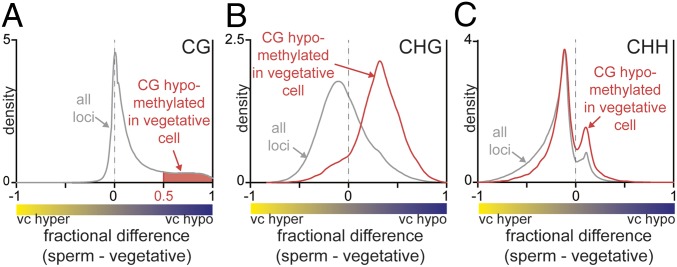
To compare sperm and vegetative cell CG methylation, we calculated the pairwise CG methylation difference within 50-bp windows by subtracting the vegetative cell DNA methylation level from the sperm DNA methylation level. The kernel density of methylation difference indicated that CG methylation is globally unchanged (peak at zero), but a significant loss of DNA methylation (positive peak) occurred locally in the vegetative cell (Fig. 1A). These differentially CG-methylated regions (named CG DMRs; SI Appendix, Methods) (10) consisted of 105,909 50-bp windows. We then determined how non-CG methylation behaves in the identified CG DMRs (shaded in red, Fig. 1A and Dataset S1). CHG methylation in CG DMRs was likewise hypomethylated (Fig. 1B, red trace), even though CHG is globally hypermethylated compared with sperm (Fig. 1B, gray trace). A weaker correspondence exists for CHH methylation (Fig. 1C), perhaps because RdDM patterns are more variable and may be partially restored after DNA demethylation. These results suggest that non-CG DNA demethylation occurs primarily in discrete regions displaying CG demethylation.
Fig. 1.
Local hypomethylation occurs in rice vegetative cells (vc). (A) Density plot showing the frequency distribution of CG methylation differences between sperm cell and vegetative cell 50-bp windows. The windows with a difference of at least 0.5 are used as vegetative cell DMRs regions compared with sperm (shaded in red, CG DMRs). (B and C) Density plots showing the frequency distribution of CHG methylation (B) and CHH methylation (C) differences between sperm and vegetative cells. The analysis marked by red is confined to CG-hypomethylated loci (shaded in A), and the gray trace contains all loci.
To determine where the CG hypomethylation occurs broadly in the genome, we used a less stringent filter to identify the hypomethylated sites in the genome as previously described (SI Appendix, Methods) (22). Also, these broad demethylated sites are useful for identifying where these hypomethylated regions are predominantly located. We identified 75,064 loci (Dataset S2, named low-stringency DMRs) that were significantly hypomethylated at CG sites in the vegetative cell compared with sperm. They are preferentially found in TEs and nonannotated regions, unlike endosperm DMRs (compared with the embryo) that were largely found in gene introns and nonannotated regions (SI Appendix, Fig. S3A) (22). These loci containing TEs were predominantly short in size (less or equal to 500 bp), even though the majority of TEs in the genome are over 5 kb in length (SI Appendix, Fig. S3B). The low-stringency DMRs (Dataset S2) contain 92% of the CG DMRs in Fig. 1A (red-shaded region). The remaining CG DMRs (8% of the total), surrounded by less demethylated sites in the vegetative cells compared with sperm, were excluded from the low-stringency DMRs in Dataset S2 (SI Appendix). Conversely, 16% of low-stringency DMRs overlap with CG DMRs. Because the low-stringency DMRs contain a large number of 50-bp windows with much less methylation difference and with low sequence coverage, we utilized the more robust CG DMRs for our further analyses.
Previous studies identified that Arabidopsis vegetative cell DMRs are located predominantly in euchromatic TEs (5). To determine whether rice vegetative cell DMRs are preferentially found in euchromatic TEs, we analyzed the correlation between the level of CG hypomethylation in TEs with CG content associated with long heterochromatic TEs, as well as the enrichment of heterochromatin marks (H3K9me2 and H3K9me1) (23). The loci depleted for heterochromatic features (SI Appendix, Fig. S4 A–D, yellow boxes) lost more DNA methylation in the vegetative cell compared with the loci that were more heterochromatic (SI Appendix, Fig. S4 A–D, blue boxes). Consistently, the sites enriched for euchromatic marks (H3K27me3, H3K4ac, H3K9ac, and H3K27ac) lost more DNA methylation (SI Appendix, Fig. S4 E–H, blue boxes) in the vegetative cell compared with the loci that were less euchromatic (SI Appendix, Fig. S4 E–H, yellow boxes). The correlation between vegetative cell hypomethylation and heterochromatin mark depletion as well as with euchromatic mark enrichment suggests that the loss of DNA methylation occurs more frequently in euchromatic TEs. This profile strongly resembles what was observed previously in Arabidopsis (10, 24), suggesting that the rice DNA demethylation glycosylase is affected by chromatin structure very similarly to the Arabidopsis DME demethylation glycosylase.
Vegetative Cell Local Hypomethylation Requires ROS1a.
A loss-of-function mutation in the rice ROS1a gene displays DME-like phenotypes such as seed abortion and low pollen germination ratio, suggesting that ROS1a functions like DME during rice seed development (18). To determine whether ROS1a has a role in DNA demethylation in pollen, especially in the vegetative cell, we isolated sperm cells and vegetative cell nuclei from heterozygous ros1a/+ plants (the strong ros1a allele cannot be made homozygous) (18), which also expressed a UBQ::H2B-GFP transgene to facilitate visualization of vegetative and sperm nuclei. Due to the previously reported Mendelian 1:1 segregation of wild-type and ros1a alleles during meiosis (18), the population of sperm and vegetative cells isolated from heterozygous ros1a/+ plants were predicted to have a 1:1 mixture of ros1a:wild-type alleles. We constructed bisulfite-treated genomic libraries using 398 sperm and 277 vegetative cell nuclei and generated maps of DNA methylation with a sevenfold genome coverage for both samples (SI Appendix, Table S1).
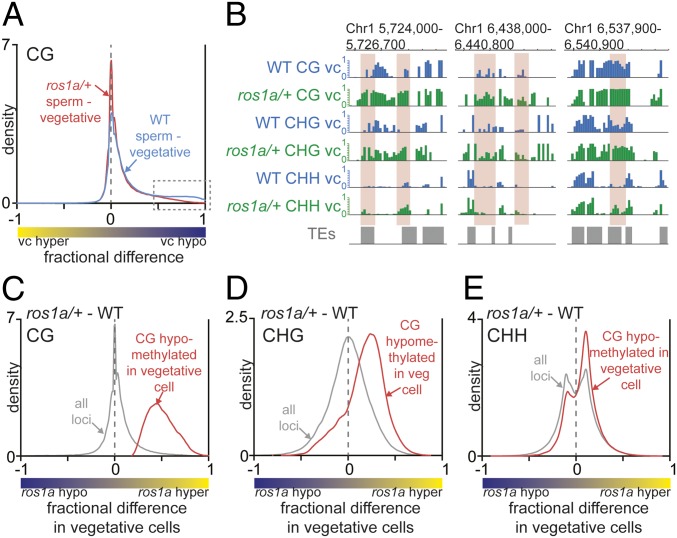
The DNA methylation profiles at TEs of vegetative cells isolated from ros1a/+ plants showed a slight global increase in CG methylation compared with wild type (SI Appendix, Fig. S2A). By contrast, there was no appreciable difference between mutant and wild-type sperm TEs (SI Appendix, Fig. S2A). Comparing sperm and vegetative cells by kernel density analysis revealed that the local hypomethylation we detected in wild-type vegetative cells versus sperm decreased in vegetative cells versus sperm cells isolated from ros1a/+ plants (Fig. 2A, dotted box). This result suggests that ROS1a-mediated DNA demethylation is responsible for local hypomethylation in the vegetative cell. To directly examine the effects of the ros1a mutation on CG DMRs, we calculated DNA methylation differences between vegetative cells isolated from wild-type and ros1a/+ plants. Globally, there was no change in CG methylation between ros1a/+ and wild type (Fig. 2C, gray trace, peak at zero), but all sequences that overlap with CG DMRs had a higher DNA methylation level in ros1a/+ compared with wild type (Fig. 2C, red trace, positive peak). Similar results were observed by directly comparing individual TEs (Fig. 2B). Moreover, the peak of fractional differences between ros1a/+ and wild type in the CG DMRs was close to 0.5 (Fig. 2C, red trace), likely because our mutant sample is heterozygous, and the data display the average DNA methylation difference between an equal mixture of ros1a and wild-type vegetative cells. Similar to CG methylation, CHG methylation in vegetative cells isolated from ros1a/+ plants also showed a higher DNA methylation level at CG DMRs (Fig. 2 B and D). A similar but weaker correspondence exists for CHH methylation (Fig. 2 B and E), presumably because sRNA-directed DNA methylation (RdDM) patterns are more variable and may be partially restored after DNA demethylation. Taken together, these results suggest that ROS1a is responsible for local DNA demethylation in rice vegetative cells and removes DNA methylation at target sites in all sequence contexts.
Fig. 2.
ROS1a is required for the local hypomethylation in the vegetative cell. (A) Density plot of CG methylation differences between sperm cell and vegetative cell 50-bp windows. (B) Snapshots of DNA methylation in wild-type and ros1a/+ vegetative cell (vc). DMRs between wild type and ros1a/+ are highlighted in red. (C–E) Density plots of CG (C), CHG (D), and CHH (E) methylation differences between ros1a/+ vegetative cells and wild-type vegetative cells. The red trace analysis is confined to CG-hypomethylated loci in wild-type vegetative cell compared with wild-type sperm, and the gray trace represents all loci from Fig. 1A.
ROS1a Indirectly Regulates Sperm Non-CG Methylation.
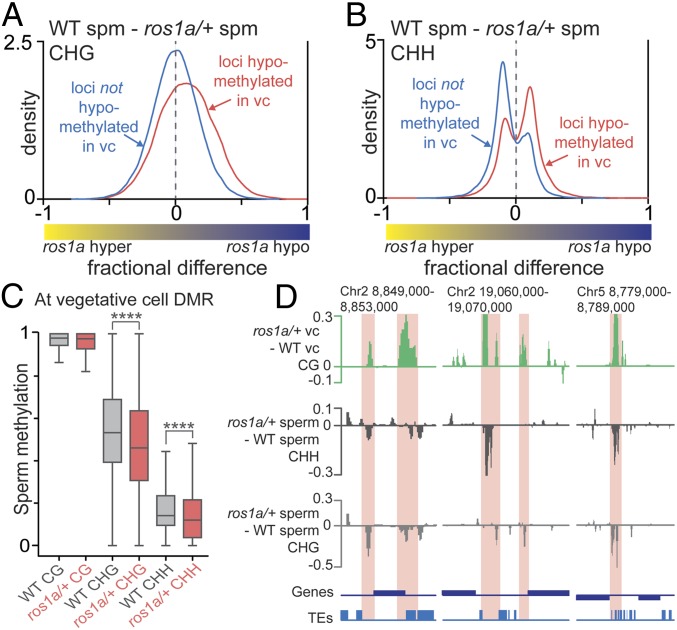
Previous studies in Arabidopsis suggested that DME DNA methylation in the vegetative cell indirectly promoted non-CG methylation in sperm cells (10). We thus set out to determine the extent to which the ros1a mutation alters sperm non-CG DNA methylation. At control loci not hypomethylated in the vegetative cell (compared with sperm), there was no detectable difference in CHG methylation in wild-type sperm compared with sperm isolated from ros1a/+ plants (Fig. 3A, blue trace peak centered at zero). By contrast, at loci hypomethylated in vegetative cells (ROS1a targets), wild-type sperm were CHG-hypermethylated compared with sperm isolated from ros1a/+ plants (Fig. 3A, red trace peak greater than zero). A similar trend was observed for CHH methylation (Fig. 3B). CG methylation, however, showed a minimal difference but with a similar trend as non-CG methylation (SI Appendix, Fig. S5). Consistent with the density plots, the absolute level of sperm DNA methylation at vegetative cell hypomethylated loci was lower in sperm isolated from ros1a/+ plants compared with wild type at non-CG sequences (Fig. 3C). Finally, ros1a/+ CG DMRs in the vegetative cell overlap sites displaying decreased CHH and CHG methylation in ros1a/+ sperm (Fig. 3D). Taken together, these results suggest that ROS1a DNA demethylation in the vegetative cell indirectly functions to promote non-CG methylation in rice sperm.
Fig. 3.
ROS1a indirectly affects non-CG methylation in sperm. (A and B) Density plots of CHG (A) and CHH (B) methylation differences between wild-type sperm and ros1a/+ sperm. The blue traces display control loci that are not hypomethylated in the vegetative cell (vc), with the sperm and vegetative cell methylation difference less than zero in CG, CHG, and CHH (see Fig. 1 A–C, gray trace). The red traces in Fig. 3 A and B contain the CG DMRs in Fig. 1A (shaded region) that also have sperm and vegetative CHG and CHH methylation differences bigger than zero, as shown in Fig. 1B (red trace) and Fig. 1C (red trace), respectively. The red trace containing loci in Fig. 3 A and B corresponded to 24% of all CG DMRs found under the red-shaded region in Fig. 1A. (C) Box plots with absolute DNA methylation levels of 50-bp windows in sperm at CG-hypomethylated loci in vegetative cells (****P < 0.0001, Wilcoxon’s matched-pairs signed-rank test). (D) Snapshots of direct and indirect target sequences of ROS1a in the vegetative cell and in the sperm, respectively. The graph was smoothed by taking the median of three consecutive 50-bp windows. The sites regulated by ROS1a in vegetative cell and sperm are highlighted in red.
ROS1a Target Sequences in the Vegetative Cell Overlap Endosperm and Central Cell DMRs.
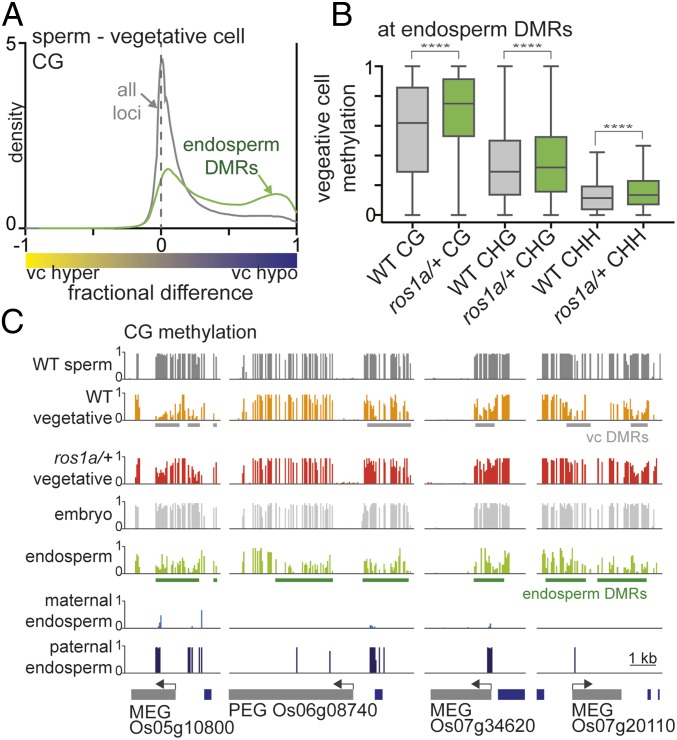
Arabidopsis DME-demethylated sequences in the vegetative cell display a 50% overlap with demethylated sequences in the maternal endosperm genome (10). Given that ROS1a is a functional analog of DME in the rice male gametophyte, we asked whether similar sequences are hypomethylated in rice vegetative cell, central cell, and endosperm. To this end, we plotted and compared vegetative cell CG DMRs (Fig. 1A, shaded in red) with previously published hypomethylated sites in the central cell compared with egg cell (central cell DMR, Dataset S3) and endosperm compared with embryo (endosperm DMR, Dataset S4) (9, 17). Vegetative cell DMRs were substantially shared with endosperm and central cell DMRs, as indicated by the larger peak on the right side of the density distribution (Fig. 4A, green trace and SI Appendix, Fig. S6A, blue trace) compared with all loci control (Fig. 4A and SI Appendix, Fig. S6A, gray traces). Vegetative cell DMRs overlapped with 40 and 32% of endosperm DMRs and central cell DMRs, respectively (SI Appendix, Fig. S6 C and D). Additionally, the absolute DNA methylation levels in the vegetative cell at endosperm DMRs (Fig. 4B) or central cell DMRs (SI Appendix, Fig. S6B), respectively, were significantly higher in ros1a/+ mutant plants compared with wild type. The overlap of vegetative cell, central cell, and endosperm DMRs, along with the genetic requirement for ROS1a for viable seed and its expression in ovules (18), is consistent with ROS1a acting as a DNA demethylating glycosylase in the central cell.
Fig. 4.
ROS1-targeted DMRs in the vegetative cell overlap with endosperm DMRs. (A) Density plots of CG methylation differences between sperm and vegetative cells. The gray trace represents all loci, and the green trace contains the sequences that overlap with hypomethylated sites in endosperm compared with embryo. (B) Box plots displaying absolute DNA methylation levels of wild-type and ros1a/+ vegetative cell 50-bp windows (****P < 0.0001, Wilcoxon’s matched-pairs signed-rank test). (C) Snapshots of ROS1a target sites near maternally expressed genes (MEG) and a paternally expressed gene (PEG). Previously published endosperm and embryo data are from 17.
ROS1 Family Proteins May Imprint Parent-of-Origin Expressed Genes.
Localized DNA demethylation in Arabidopsis and rice endosperm is inherited maternally from the central cell and is crucial for parent-of-origin–specific, imprinted gene expression (9). Some Arabidopsis-imprinted genes are also demethylated and expressed specifically in the vegetative cell of pollen (7). Given that vegetative cell DMRs and endosperm DMRs overlap significantly in rice, we determined whether ROS1a target sequences in the vegetative cell are also located near endosperm-imprinted genes. We determined that vegetative cell DMRs mediated by ROS1a were near previously identified imprinted genes [Fig. 4C, maternally expressed genes (MEG) and paternally expressed genes (PEG)] that also displayed differential methylation of maternal and paternal alleles (22). We identified 45 endosperm-imprinted genes with at least twofold increased expression in vegetative cells compared with sperm cells (25). Of those 45 genes, 19 genes (42%) showed DNA hypomethylation in both endosperm and vegetative cell compared with egg and sperm, respectively (SI Appendix, Fig. S7A, orange and light gray slices), and ROS1a was required for hypomethylation at 16 of these 19 genes (84%) (SI Appendix, Fig. S7A, orange slice). Moreover, previously identified genes with parent-of-origin expression that affect seed size or composition (Os07g20110, Os01g08570, Os06g30280, Os10g37540, and Os04g08034) (26) displayed ROS1-mediated hypomethylation in the vegetative cell (SI Appendix, Fig. S7B). These results are consistent with the ROS1a protein and/or possibly other ROS1 family proteins regulating gene expression by establishing gene imprinting in the central cell and endosperm.
Global CG Methylation Levels Are Elevated in Rice Male Gametes and Companion Cells.
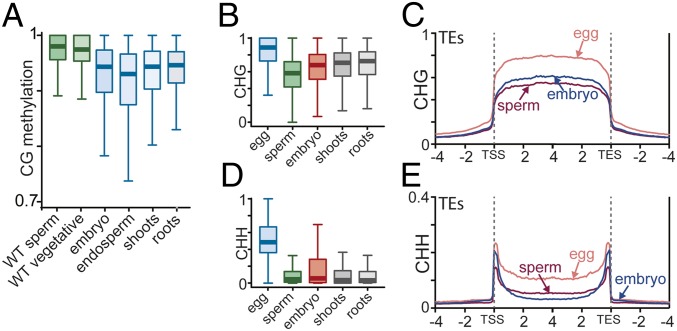
CG methylation is globally higher in the Arabidopsis male sexual lineage (vegetative cells, sperm cells, and the microspores from which they arise) and in rice female gametes (egg and central cells) compared with somatic tissues (leaf and root), which may reflect a more efficient mechanism for maintaining CG methylation in sexual lineages (9, 24). Therefore, we asked whether rice sperm and vegetative cell CG methylation is also globally elevated compared with somatic tissues outside of the local hypomethylated regions. Similar to Arabidopsis, rice sperm and vegetative cells displayed elevated levels of TE CG methylation compared with endosperm, embryo, and other somatic tissues (Fig. 5A). These results suggest that CG methylation is more efficiently maintained in the rice male sexual lineage compared with somatic tissues.
Fig. 5.
DNA methylation reprogramming occurs during embryogenesis. (A) Box plots showing DNA methylation levels of 50-bp windows in TEs of the indicated cell or tissue type. Only sites outside DMRs in the CG context between rice endosperm and embryo are included in the analysis. (B and D) Box plots showing DNA methylation levels of 50-bp windows in TEs of the indicated cell or tissue type. (C and E) TEs were aligned at the 5′ (TE start site, TSS) and 3′ (TE end site, TES) ends. CHG (C) and CHH (E) methylation within each 100-bp interval were averaged and plotted from 4 kb away from the annotated TE (negative numbers) to 4 kb into the annotated TE (positive numbers). The dashed lines represent the points of alignment.
Sperm and Central Cell Genomes Are Globally Hypomethylated in the Endosperm.
In Arabidopsis and rice endosperm, global CG hypomethylation was observed previously (17, 24). Since the CG methylation of the Arabidopsis and rice central cell, the origin of maternal genomes inherited in the endosperm, is globally elevated (9), endosperm global CG hypomethylation must be established after fertilization. However, the methylation status of sperm, the endosperm’s paternal-inherited genome, is unknown in rice. To address this question, we compared DNA methylation of rice sperm, central cell, and endosperm in genes and TEs. We found that sperm CG methylation in TEs and genes was elevated compared with endosperm (SI Appendix, Fig. S8 A and D). In addition to CG methylation, CHG (SI Appendix, Fig. S8 B and E) and CHH (SI Appendix, Fig. S8 C and F) methylation in sperm and central cell also showed global hypermethylation both in genes and TEs compared with the endosperm. These results suggest that there is likely to be reduced activity of the distinct DNA methylation pathways for CG, CHG, and CHH methylation to achieve global passive hypomethylation in the endosperm after fertilization.
Non-CG Methylation of Maternal and Paternal Genomes Is Globally Reprogramed After Fertilization.
Recent studies identified that global DNA methylation reprogramming in plant embryo occurs after fertilization (27, 28); however, how DNA methylation of egg and sperm genomes transforms after fertilization is largely unknown. To address this question, we compared the DNA methylation status of sperm, egg cell, and embryo. In embryo, CG methylation level was slightly reduced in TEs compared with egg (SI Appendix, Fig. S9A) and sperm (Fig. 5A and SI Appendix, Fig. S9A). In genes, CG methylation levels are essentially identical between gametes and embryo (SI Appendix, Fig. S9B). CHG methylation, however, was much higher in egg in both TEs and genes, but was lower in sperm compared with embryo in TEs (SI Appendix, Fig. S9C and Fig. 5 B and C). Also, the absolute level of CHG methylation difference was very apparent between egg and sperm (Fig. 5B). By contrast, embryo, shoot, and roots showed similar level of CHG methylation (Fig. 5B). We then determined whether these strikingly different CHG methylation levels of maternal-derived and paternal-derived genomes are maintained after fertilization in the embryo. To compare the genomes before and after fertilization, we computationally separated maternal and paternal genomes of the hybrid embryo using single-nucleotide polymorphism (SNP) differences as described previously (22). Unlike the gametes, the maternal and paternal embryo genomes showed similar levels of CHG methylation (SI Appendix, Fig. S10A, red boxes), suggesting that CHG DNA methylation on the maternal and paternal alleles equilibrates after fertilization. Similarly, the egg CHH methylation level is much higher (Fig. 5D and SI Appendix, Fig. S10B, blue box) compared with sperm as well as maternal-derived embryo CHH methylation (Fig. 5D and SI Appendix, Fig. S10B, green and red boxes). By contrast, the median of sperm CHH methylation level (Fig. 5D, green box) was very similar to the median of paternal-derived embryo (Fig. 5D, red box). Thus, there is a dramatic equilibration of CHH DNA methylation at the maternal- and paternal-derived genomes after fertilization.
We then studied CHH methylation more carefully by plotting CHH methylation levels of sperm, egg, and embryo along the length of TEs. We found that egg CHH methylation was much higher compared with sperm and embryo, consistent with our box plots (Fig. 5E). The sperm CHH methylation, however, was lower compared with embryo near the TE points of alignment (Fig. 5E) that are enriched for short TEs and edges of long TEs, which tend to be euchromatic and sometimes reside at the promoter of genes (SI Appendix, Fig. S9D) (5, 17). By contrast, the long, autonomous heterochromatic TEs (SI Appendix, Fig. S9E) enriched within the TE and away from the TE points of alignment (Fig. 5E), showed the opposite CHH methylation phenotype where CHH methylation was higher in sperm compared with embryo. These results suggest that RdDM (mostly maintaining CHH methylation at short euchromatic TEs and at edges of long TEs) and CMT2 (mostly maintaining CHH methylation within long heterochromatic TEs) are distinctly regulated during embryogenesis (5). Overall, our comparisons of non-CG methylation before fertilization (egg cells and sperm cells) and after fertilization (embryo) suggest that a complex non-CG methylation reprogramming occurs after fertilization and equilibrates egg- and sperm-derived genomes in embryo.
Discussion
Active DNA demethylation by DNA glycosylases is shown to play a key role in seed development in plants by regulating DNA methylation patterns in male and female gametophytes. In Arabidopsis, DME is responsible for active DNA demethylation in the central cell and the vegetative cell (7, 9, 10). In rice, no DME ortholog has been detected; instead, multiple ROS1 orthologs were identified using DNA sequence similarity (17). However, genetic (18) and genome-wide DNA methylation analyses (Figs. 1 and 2) show that ROS1a activity results in DNA demethylation in the vegetative cell of rice, similar to DME in Arabidopsis. Additionally, ROS1a promoter activity is also observed in the rice female sexual lineage similar to DME (18), suggesting a role of ROS1a in establishing endosperm hypomethylation (17). Our data indicate that active DNA demethylation by glycosylases in pollen is conserved in two major plant branches, which are separated by 150 million years of evolution (19).
In Arabidopsis, DME-mediated hypomethylated regions overlap significantly between central cell and maternal endosperm (a proxy for DME activity in the central cell) and the vegetative cell, suggesting an overlapping role of DME in these two different cell types (7–10, 29). Similarly, in rice, ROS1a is expressed in both the pollen and ovules (18). Also, we determined a significant overlap of the DMRs between vegetative cell and sperm (vegetative cell DMRs) and the DMRs between central cell and egg (central cell DMRs) (Fig. 4A and SI Appendix, Fig. S6A). This suggests that not only does ROS1a target methylated sequences in the vegetative cell, but also may target sites in the central cell, which program gene imprinting in the endosperm (Fig. 4C). The ratio of overlapping sequences between sperm versus central cell and endosperm was not 100%; instead, it was 32 and 40%, respectively (SI Appendix, Fig. S6 C and D). This is also reminiscent of what we have seen in Arabidopsis, whereby vegetative cell and endosperm DMRs overlap by ∼50% (10). The nonoverlapping DMRs between the vegetative cell and central cell in rice could be due to the differences in chromatin structure between these two cell types, which is presumably important for recruiting ROS1a to the target sequences (Fig. 2). Moreover, the presence of other ROS1a family proteins (ROS1c, ROS1d, and DML3a) in rice ovule may be responsible for central cell and endosperm DNA demethylation. A previous study identified that other ROS1a family proteins are moderately expressed compared with ROS1a in female but not in the male gametophyte (18). It is possible that other ROS1a family proteins have different targeting preferences than ROS1a, potentially contributing to differences in demethylated sites between the rice central cell and endosperm, as well as the vegetative cell.
Our data show that ROS1a-mediated DNA demethylation reinforces sperm non-CG DNA methylation (Fig. 3). One possibility is that there is communication between sperm and vegetative cells and that the signal is generated by ROS1a-mediated DNA demethylation in the vegetative cells. The signal could be part of the RdDM pathway because it targets non-CG DNA methylation at euchromatic TE sequences (5). The signal could be an mRNA fragment or a small RNA molecule, as it has been shown that RNA can move from the vegetative cell to sperm cell (30, 31). Consistent with this idea, it has been proposed that sRNA, generated by active DNA demethylation in the vegetative cell, is responsible for DNA methylation reprogramming in the sperm cells of Arabidopsis (10, 16). This reinforcement of DNA methylation by the vegetative cell also highlights another role of vegetative cells, where active DNA demethylation in this cell type, potentially resulting in harmful transposition of mobile elements, may be used to generate sRNAs that mediate DNA methylation to immunize the sperm and, therefore, the next generation against TE activation.
In animals, global DNA methylation reprogramming in embryo occurs after fertilization (32). Our data show that global DNA methylation reprogramming occurs in both rice endosperm and embryo, the tissues from double-fertilization in plants. In rice endosperm, global CG and non-CG DNA methylation are reduced after fertilization both in TEs and genes (SI Appendix, Fig. S8), suggesting that all maintenance DNA methyltransferases, MET1, CMT3, and CMT2, are less active during endosperm development contributing to globally passive CG and non-CG hypomethylation. Consistent with our results, MET1 in Arabidopsis endosperm is significantly down-regulated compared with the embryo, and the residual CG methylation may be catalyzed by other distinct MET family members (33).
Dramatic reprogramming of non-CG methylation has also been observed in Arabidopsis root (27) and embryo development (28, 34). DNA methylation reprogramming also occurs in the rice embryo after fertilization, as shown by comparing embryo-specific DNA methylation patterns with sperm and egg methylation patterns (Fig. 5 C and E and SI Appendix, Fig. S10). Notably, rice sperm and egg cells have significantly different global non-CG methylation levels (Fig. 5). These dissimilar levels of DNA methylation of male and female gametes are also found in mammals, fish, and insects (35–37). In mammals, the global percentage of CG methylation in sperm is much higher than that of the oocyte. These very different levels of gamete DNA methylation in animals, however, equilibrate after fertilization, where both maternal and paternal genomes undergo active demethylation in the zygote before embryogenesis (35). In rice, we observed that global non-CG methylation levels are very different in sperm and egg cells, but their DNA methylation levels are similarly equilibrated in the embryo (SI Appendix, Fig. S10), alluding to potential DNA reprogramming during embryogenesis in plants.
Materials and Methods
Complete details concerning plant materials, experimental methods, and data analyses are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Jennifer Frost for helpful discussions, Shigeru Iida for providing the ros1a/+ mutant line, and Ms. T. Mochizuki for isolating pollen grains. This work was funded by NIFA-USDA (2013-67012-21112) Postdoctoral Fellowship (to M.Y.K.); NSF (IOS-1025890) and NIH (R01-GM069415) grants (to R.L.F. and D.Z.); Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (JP15K07262) (to A.O.); Ministry of Education, Culture, Sports, Technology and Science (MEXT) Grant-in-Aid for Scientific Research (JP16H06471) (to T.K.); and MEXT Grant-in-Aid for Scientific Research (17H058452) and JSPS Grant-in-Aid for Scientific Research (16K14742) (to T.O.). The University of California, Berkeley, Vincent J. Coates Genomics Sequencing Laboratory is supported by an NIH grant (S10 OD018174).
Footnotes
The authors declare no conflict of interest.
Data deposition: DNA sequencing data are deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE126791).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821435116/-/DCSupplemental.
References
- 1.Awika JM. 2011. Major cereal grains production and use around the world. Advances in Cereal Science: Implications to Food Processing and Health Promotion, ACS Symposium Series, eds Awika JM, Piironen V, Bean S (American Chemical Society, Washington, DC), Vol 1089, Chap 1.
- 2.Cantrell RP, Reeves TG. The rice genome. The cereal of the world’s poor takes center stage. Science. 2002;296:53. doi: 10.1126/science.1070721. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J-K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108:8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 9.Park K, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA. 2016;113:15138–15143. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J-S, et al. Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2017;114:2078–2083. doi: 10.1073/pnas.1620592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost JM, et al. FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Proc Natl Acad Sci USA. 2018;115:E4720–E4729. doi: 10.1073/pnas.1713333115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satyaki PRV, Gehring M. DNA methylation and imprinting in plants: Machinery and mechanisms. Crit Rev Biochem Mol Biol. 2017;52:163–175. doi: 10.1080/10409238.2017.1279119. [DOI] [PubMed] [Google Scholar]
- 14.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang K, Lang Z, Zhang H, Zhu J-K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat Plants. 2016;2:16169. doi: 10.1038/nplants.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ono A, et al. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012;71:564–574. doi: 10.1111/j.1365-313X.2012.05009.x. [DOI] [PubMed] [Google Scholar]
- 19.Chaw S-M, Chang C-C, Chen H-L, Li W-H. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 20.Toda E, Okamoto T. Formation of triploid plants via possible polyspermy. Plant Signal Behav. 2016;11:e1218107. doi: 10.1080/15592324.2016.1218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smallwood SA, Kelsey G. De novo DNA methylation: A germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues JA, et al. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci USA. 2013;110:7934–7939. doi: 10.1073/pnas.1306164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Y, et al. Histone modifications facilitate the coexpression of bidirectional promoters in rice. BMC Genomics. 2016;17:768. doi: 10.1186/s12864-016-3125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh P-H, et al. Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc Natl Acad Sci USA. 2016;113:15132–15137. doi: 10.1073/pnas.1619074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson SN, et al. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: Evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 2013;76:729–741. doi: 10.1111/tpj.12336. [DOI] [PubMed] [Google Scholar]
- 26.Yuan J, et al. Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol. 2017;216:373–387. doi: 10.1111/nph.14510. [DOI] [PubMed] [Google Scholar]
- 27.Kawakatsu T, Nery JR, Castanon R, Ecker JR. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017;18:171. doi: 10.1186/s13059-017-1251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouyer D, et al. DNA methylation dynamics during early plant life. Genome Biol. 2017;18:179. doi: 10.1186/s13059-017-1313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, et al. Intercellular communication in Arabidopsis thaliana pollen discovered via AHG3 transcript movement from the vegetative cell to sperm. Proc Natl Acad Sci USA. 2015;112:13378–13383. doi: 10.1073/pnas.1510854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez G, Panda K, Köhler C, Slotkin RK. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants. 2016;2:16030. doi: 10.1038/nplants.2016.30. [DOI] [PubMed] [Google Scholar]
- 32.Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh T-F, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108:1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J-Y, et al. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc Natl Acad Sci USA. 2017;114:E9730–E9739. doi: 10.1073/pnas.1716758114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 36.Jiang L, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drewell RA, et al. The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development. 2014;141:2702–2711. doi: 10.1242/dev.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.