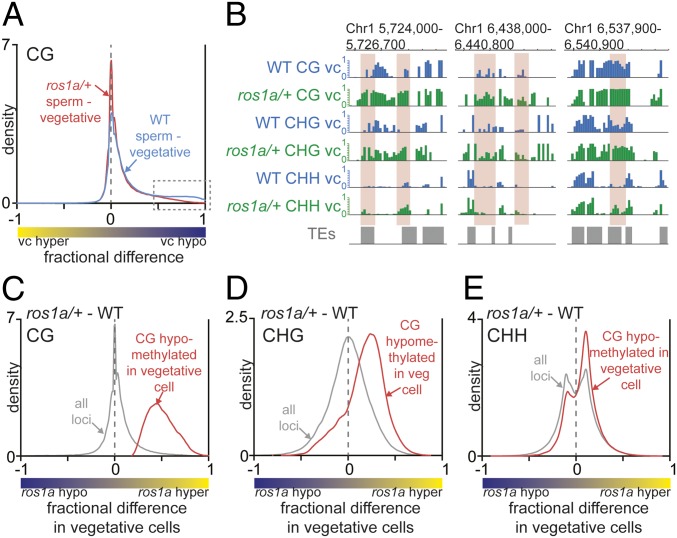
Fig. 2.
ROS1a is required for the local hypomethylation in the vegetative cell. (A) Density plot of CG methylation differences between sperm cell and vegetative cell 50-bp windows. (B) Snapshots of DNA methylation in wild-type and ros1a/+ vegetative cell (vc). DMRs between wild type and ros1a/+ are highlighted in red. (C–E) Density plots of CG (C), CHG (D), and CHH (E) methylation differences between ros1a/+ vegetative cells and wild-type vegetative cells. The red trace analysis is confined to CG-hypomethylated loci in wild-type vegetative cell compared with wild-type sperm, and the gray trace represents all loci from Fig. 1A.