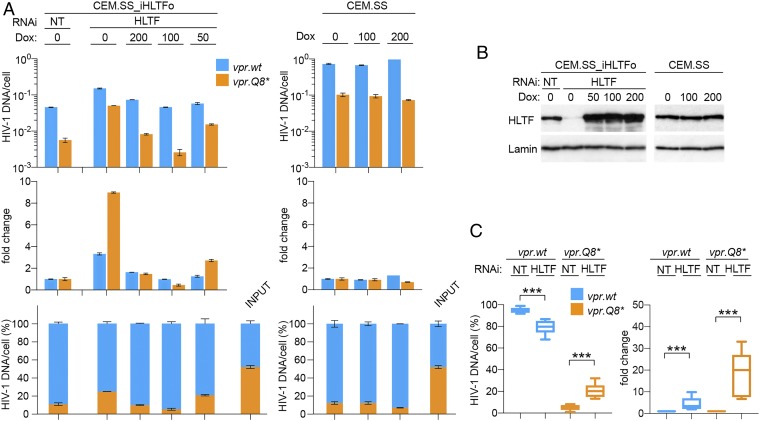
Fig. 3.
HIV-1 Vpr antagonizes HLTF-mediated restriction of HIV-1 replication in T cells. (A) HLTF restricts HIV-1 replication. CEM.SS T cells harboring doxycycline-inducible codon-optimized HLTF transgene (CEM.SS_iHLTFo) were subjected to NT or targeting endogenous HLTF mRNA (HLTF) RNAi in the absence or presence of the indicated concentrations of doxycycline (ng/mL). (Left) Three days after initiation of RNAi, PRCA was performed with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8*. (Right) As a control, PRCA was also performed in parental CEM.SS T cells cultured in the absence or presence of doxycycline. (Upper) Cell-associated DNA of the competing viruses was quantified at 7 dpi. (Middle) Fold-change in HIV-1 DNA copies in cells subjected to HLTF RNAi, normalized to those in cells subjected to NT RNAi. (Lower) Percentages of cell-associated HIV-1 DNA for the competing viruses. (B) HLTF levels in CEM.SS_iHLTFo and parental CEM.SS T cell populations used in experiments shown in A were revealed by immunoblotting. Lamin B1 provided a loading control. (C) HLTF depletion partially restores the HIV-1 replication fitness advantage provided by Vpr. CEM.SS T cells were subjected to NT or HLTF-targeting (HLTF) RNAi and infected with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8*. Percentages of cell-associated DNA for the competing viruses at 7 dpi (Left) and fold-change in HIV-1 DNA copies per cell in cells subjected to HLTF-targeting RNAi normalized to those in cells subjected to NT RNAi (Right) are shown. The data shown are from five biological replicate experiments. ***P < 0.001.