Abstract
Hepatocellular carcinoma (HCC) patients are at high risk for both local recurrence and distant metastasis and tightly associated with poor prognosis. Exploring the molecular mechanism will provide a new opportunity in developing personal treatment for advanced HCC patients. As a critical member of the Retinal Determination Gene Network (RDGN), EYA1 has been identified as a tumor promoter in various cancers; however, its role in HCC has never been investigated. The present study was aimed to explore the role of EYA1 in HCC development. By analyzing public microarray datasets, we found that the EYA1 mRNA level was enhanced in HCC, which was significantly correlated with an aggressive phenotype and poor prognosis. Besides, EYA1 was coordinated with the fibronectin type III domain containing 3B (FNDC3B) to promote the migration and invasion of HCC cells. Western blot assays indicated that EYA1 not only increased the abundance of FNDC3B but also contributed to Epithelial-Mesenchymal Transition (EMT)-like phenotype change, like increased N-cadherin and decreased E-cadherin expression. Collectively, this study suggested that EYA1 activated FNDC3B to promote the migration and invasion in HCC. The aberrant expressions of EYA1 and FNDC3B may become the poor predictors for HCC patients.
Keywords: EYA1, FNDC3B, hepatocellular carcinoma, EMT, tumor growth
Introduction
Hepatocellular carcinoma (HCC) is the sixth common malignancy and the third leading cause of cancer mortality worldwide, with more than 780,000 cases diagnosed and 740,000 deaths annually [1]. Although there is impressive progress in combination therapy for HCC patients, like the success of lenvatinib in the frontline and regorafenib in the second line chemotherapy, the prognosis remains unfavorable. The five-year survival rate of the HCC patients is less than 30% [2,3]. The development of HCC is involved in the accumulation of genetic and epigenetic mutations of particular oncogenes. Aberrant activation of specific signaling pathways might function as a critical regulator in hepatocarcinogenesis and metastasis [4]. Therefore, understanding the molecular basis of HCC is essential for developing effective strategies for this malignancy.
Eya belongs to the retinal determination gene network (RDGN) family, which also contains a dominant suppressor dachshund (dac/Dach) and the Six family transcription factor sine oculis (so/Six) [5]. The EYA family is homologous to the Drosophila eyes absent (eya) gene, a key regulator for Drosophila eye morphogenesis [6]. In vertebrates, there are four encode Eya proteins (Eya1-4). They are characterized by a 271-amino acid carboxyl-terminal motif named EYA domain (ED), which is conserved between species and exhibits dual phosphatase activities [3,7]. Another conserved EYA domain/N-terminal domain (ED2/NTD) contains a rich stretch of proline/serine/threonine residues and shows Ser/Thr phosphatase activity [5]. In humans, mutations in EYAs or disruption of the SIX/EYA complex can cause branchio-oto-rena (BOR) syndrome, an autosomal dominant genetic disorder marked by undeveloped or absent kidneys, deafness, auricular malformations and bronchial arch remnants [8-10]. EYA family members keep silenced when cell, tissue and organ finish the differentiation [6]. However, abnormal re-activation of embryo development related genes can trigger the tumorigenesis and progression. Over the past decade, the overexpression of EYA family members has become one of “hallmarks of cancer”, including sustained proliferative signaling, resistance to cell death, angiogenesis, invasion, and metastasis [11-13].
Basically, EYA family has two roles, either shows the capacity to phosphorylate on tyrosine and threonine, or functions as the essential transcriptional cofactors for the SIX family of homeoproteins [14]. EYA’s Tyr phosphatase activity have a broad effect on transcriptional regulation [15], survival response to DNA damage by de-phosphorylating H2AX [12], and inhibition of the anti-tumor activity of ERβ [16]. Among them, the role of EYA1 in tumorigenesis and tumor development has attracted wide attention. Overexpression of EYA1 has been found in various types of cancer, including Wilms’ tumors [17], breast cancer [18], gastric carcinoma [19] and a subset of leukemia patients [20]. In breast cancer, exogenous expression of EYA1 contributed to breast tumor growth and induced cancer stem cell (CSCs) properties via activation of cyclin D1 [18]. On the other hand, knockdown EYA1 in breast cancer cells destabilized the Myc expression, leading to cell cycle arrest [21]. However, whether the oncogenic role of EYA1 depending on tissue type has continued to attract great interest. To date, the potential role of EYA1 in HCC has not been evaluated. In the current study, we conducted a combined analysis of available published microarray data and immunohistochemistry analysis on HCC patients’ tissues. Furthermore, the functional role of EYA1 in HCC cell lines was also investigated.
Materials and methods
Collection of tissue specimens
Primary HCC and the adjacent non-tumor (NT) liver specimens were obtained from 20 HCC patients who underwent the curative resection in Zhongnan Hospital of Wuhan University. None of them has ever received pre-operative chemotherapy or radiation therapy. The paired tissue specimens (tumor and adjacent normal tissues) were collected from the patients after informed consent, the histological confirmations were obtained from the experienced pathologists. The study protocol was in line with the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Hospital’s Protection of Human Subjects Committee.
RNA isolation, cDNA synthesis, and quantitative real-time PCR
RNA was prepared from 100 mg tissue specimens with TRIzol reagent (Invitrogen, USA). cDNA was reversed from 2 μg RNA using a reverse transcription kit (TOYOBO, Japan). RT-qPCR was performed with the SYBR® Green Real-time PCR Master Mix Kit (TOYOBO, Japan). Gene expression was normalized to GAPDH. The sequence of primers was listed as following: EYA1: (forward) 5’-GTTCATCTGGGACTTGGA-3’, (reverse) 5’-GCTTAGGTCCTGTCCGTT-3’; GAPDH: (forward) 5’-CAATGACCCCTTCATTGACC-3’, (reverse) 5’-GATCTCGCTCCTGGAAGATG-3’.
Immunohistochemistry (IHC) staining and quantification analysis
The fixed specimens were embedded in paraffin and cut into 4-5 μm thick sections. IHC staining was performed as previously described [22]. Whole slide image was captured by EVOS auto cell image system (Life technology, USA). The immunohistochemical scores were assessed by two experienced pathologists without knowledge of patients’ information. For quantification, all stains were assessed at 200× magnifications, and at least 3 fields from each core were counted. Scores were determined by the intensity and percentage of positive staining tumor cell nuclei or cytoplasm in the whole tissue stains as previously described [22,23].
Cells culture and transfection
Human hepatocellular carcinoma cell lines Huh7, SMMC-7721, Hep3B and HepG2 were cultured in the recommended condition. Hep3B cells were seeded at 50% confluence on the day before transfection. HEK 293T cells were transfected with either control vector or pcDNA3.1-EYA1 expression plasmid using Lipofectamine™ 2000 (Invitrogen, Carlsbad CA, USA). The supernatant was collected for further infection. After selection by puromycin, the cells with stable transfection were used for further experiments. EYA1 expression was verified by quantitative reverse transcription-PCR (qRT-PCR) and Western blot. HepG2 cells were seeded and transfected with either siRNA or scramble control (Ribobio Company, Guangzhou, China) duplexes using Lipofectamine™ 2000. The sequence of EYA1 siRNA: (sense) 5’-CAGGAAAUAAUUCACUCACAAdTdT-3’; (antisense) 5’-UUGUGAGUGAAUUAUUUCCUGdTdT-3’. About 20 nM siRNA was used per well and cells were incubated for another 48 to 72 hours before being harvested for qRT-PCR and immunoblot analysis.
Western blot analysis
Cell and tissue lysates were extracted using ice-cold RIPA buffer and measured using a bicinchoninic acid (BCA) protein assay kit (Promoter, China). Proteins were resolved in 10% SDS-polyacrylamide gels, and then blotted onto PVDF transfer membrane. The membranes were subsequently incubated with primary antibodies that targeted to EYA1 (Proteintech, China), FNDC3B (Proteintech, China), E-cadherin (Proteintech, China), N-cadherin (Cell Signaling Technology, USA), GAPDH (Cell Signaling Technology, USA) and VINCULIN (Sigma, USA) at 4°C for overnight. The membranes were subsequently incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody. The band images were digitally captured and quantified using the enhanced chemiluminescence (Kodak Image Station 4000 MM Pro, USA).
Scratch assay
To evaluate cell migration ability, 1×105 cells were seeded into 24-well plate. The cell layers were scratched to form a cell-free straight line using a 10 μl plastic pipette tip and cultured in serum free medium. The images were taken after 36 h.
Transwell invasion assay
Transwell chambers (pore size 8.0 µm) (Corning Inc., USA) were coated with Matrigel (BD Biosciences, USA). Invasion assay was performed as previously described [23]. All experiments were conducted in triplicate.
Immunofluorescent staining
Cells were seeded on 12-mm coverslips in a 24-well plate (1×105 cells/well) at 37°C for overnight. Cells were fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 and blocked using 5% goat serum for 30 min. Cells were further incubated with primary antibodies. Next day, cells were incubated with secondary antibodies for 1 h at room temperature. Nuclei were visualized by 4’,6-diamidino-2-phenylindole (DAPI). The stained cells were examined with EVOS cell image system. Blinded membranous E-Cadherin quantification was quantified by the ratio between the number of the cells with membranous E-Cadherin and the total number of DAPI-stained cells in the field of vision.
Bioinformatic analysis
The gene expression data and clinical information were downloaded from TCGA database (https://cancergenome.nih.gov/) and Gene Expression Omnibus (GEO) databases (https://www.ncbi.nlm.nih.gov/geo/). The data were applied to evaluate the association between the target genes and the clinical information like histological stage and survival time.
Statistical analysis
GraphPad Prism 6.0 software (GraphPad Software, Inc., USA) was used for the statistical analyses. The IHC scores were presented as median ± range and were tested whether the data matched normal distribution or not. If it was, then the difference between groups were conducted by using parametric statistics (Student t test), otherwise performing nonparametric statistics (Mann-Whitney test). The correlations between clinicopathological and immunohistochemical variables were calculated according to Person χ2 test. The gene expression data from public databases were presented as median ± range. The data of PCR, scratch assay, invasion assays and immunofluorescent stain assays were expressed as mean ± standard deviation (SD). Between those groups (PCR, scratch, invasion and IF assays) comparisons were conducted with Student t test. Survival curves were estimated by the Kaplan-Meier method and compared using the log-rank test or the Wilcoxon test. P values < 0.05 were considered statistically significant.
Results
High expressions of EYA1 were correlated with the progression of hepatocellular carcinoma patients
To identify the expression of EYA1 in human hepatocellular carcinoma, we employed the Cancer Genome Atlas (TCGA) database, which contained the mRNA expression profiles with the pathological characteristics as well as survival information from 374 HCC patients. This database was also included the mRNA information of 50 normal liver tissues. It was found that EYA1 was significantly upregulated in the HCC compared with the normal liver tissues (Figure 1A). We also evaluated the mRNA expression of EYA1 among 20 pairs of HCC samples with matched adjacent normal tissues by RT-qPCR analysis. In comparison with adjacent liver tissue, the expression of EYA1 was significantly increased in the tumor samples (Figure 1B), which supports the results from transcriptome sequence in TCGA datasets. To identify whether EYA1 was correlated with the development of HCC, we further analyzed the relationship between the expression of EYA1 and clinicopathological features in the TCGA cohort. The mRNA expression of EYA1 was significantly increased when the tumor progressed to an advanced stage (Figure 1C). The median survival time for patients in the high-EYA1 group was significantly shorter than the low-EYA1 group (Figure 1D). These data are indicated higher EYA1 expression can predict poor clinical outcomes of HCC patients.
Figure 1.
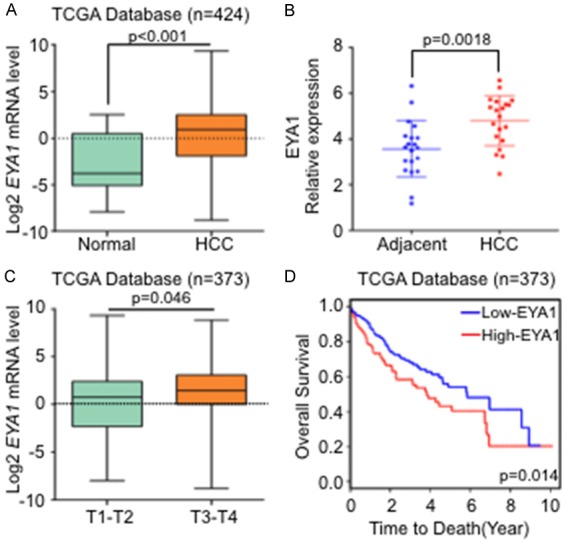
High expressions of EYA1 were correlated with the progression of hepatocellular carcinoma patients. A: TCGA database analysis showed that the mRNA expression of EYA1 in normal and HCC tissue. B: qPCR detected the EYA1 mRNA level in HCC and the adjacent liver tissues (n = 20). C: Histogram showed the EYA1 mRNA expression in T1-2 and T3-4 stage in the TCGA cohort (log2 transformed, n = 373). D: Kaplan-Meier survival curves of EYA1 in HCC based on TCGA cohort was analyzed.
EYA1 was significantly associated with FNDC3B in human HCC
To screen the potential downstream regulators of EYA1 during HCC development, we selected one dataset from Oncomine contained normal and HCC tissue [24]. After analyzing the transcriptional levels of EYA1, we ranked a cluster of genes which showed a strong correlation with EYA1 and found a novel target gene, named fibronectin type III domain containing 3B (FNDC3B) (Figure 2A). FNDC3B, also called FAD104 (factor for adipocyte differentiation 104), is a known regulator of adipocyte and osteoblast differentiation and plays a critical role in cell adhesion, growth, and migration [25,26]. In hepatocellular carcinoma, FNDC3B can promote cell migration and metastasis [27]. To verify the association between EYA1 and FNDC3B in HCC, we utilized another GEO database GSE77314. The correlation analysis indicated that EYA1 was positively associated with FNDC3B (Figure 2B). Since EYA1 may cooperate with FNDC3B in the regulation of HCC, the combined evaluation of the two genes would provide more precise information in prediction of the clinical outcome. As a result, high EYA1 and FNDC3B expression were significantly associated with poor prognosis. Patients characterized by low levels of both EYA1 and FNDC3B had the longest overall survival in this cohort (Figure 2C). Meanwhile, GSE77314 (one cohort; n = 100) or GSE14520 (two cohorts; n = 43, n = 445, respectively) were applied to detect the profiles of FNDC3B in HCC. As a result, it was suggested in comparison with normal liver tissues, the mRNA of FNDC3B in HCC was significantly increased (Figure 2D and 2E). Together, it suggests both of EYA1 and FNDC3B are increased in HCC and associated with poor prognosis.
Figure 2.
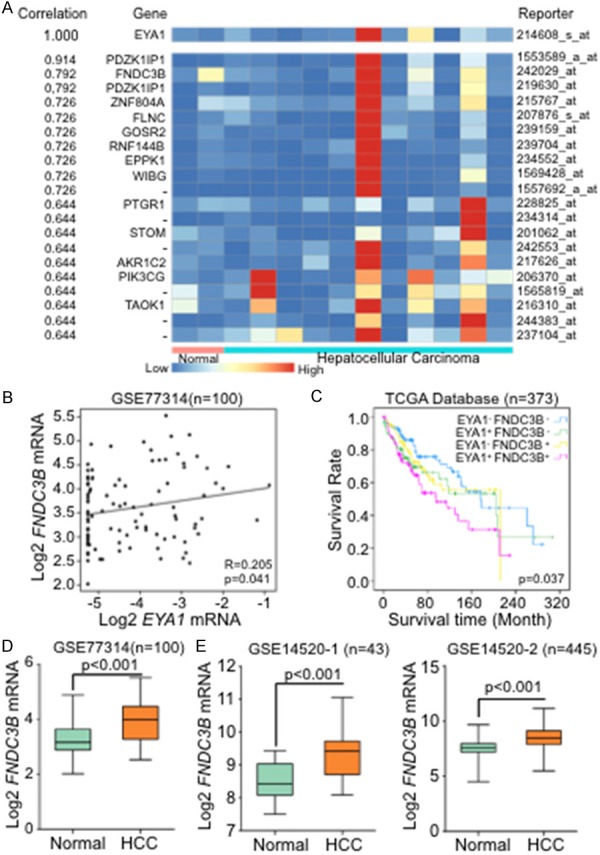
FNDC3B was identified as a downstream target of EYA1 and increased in HCC. A: Heat map based on Oncomine database analysis showed the co-expression genes of EYA1. B: The correlation analysis examined the relationship between EYA1 with FNDC3B by using a GEO database (GSE77314). C: Kaplan-Meier survival curve of EYA1 combined with FNDC3B was analyzed based on TCGA database. D and E: Analysis from GEO databases (GSE77314 and GSE14520) showed that FNDC3B mRNA expression in normal and HCC tissue.
To further identify the protein abundance of EYA1 and FNDC3B in human hepatocellular carcinoma, we detected the expressions of EYA1 and FNDC3B in 20 pairs of HCC and adjacent normal tissue via immunohistochemical (IHC) staining. Representative images of IHC staining of EYA1 and FNDC3B were shown in Figure 3A. Statistical analysis from IHC score was revealed EYA1 and FNDC3B protein abundance were significantly increased in HCC, whereas kept low expression in adjacent normal tissue (Figure 3B and 3C). Additionally, we exanimated the correlation between EYA1 and FNDC3B. Based on the IHC score of EYA1 expression, all the samples were classified as high (IHC score ≥ 6, n = 11) and low (IHC score < 6, n = 9) group. Representative images of FNDC3B staining in low and high EYA1 group were shown in Figure 3D. The semi-quantitative result indicated that FNDC3B was significantly increased in high EYA1 cases (Figure 3E). We also extracted the protein from 6 paired of HCC and adjacent tissue from this cohort. The results from western blot were consistent with IHC staining, indicating EYA1 integrated with FNDC3B and highly expressed in HCC compared with normal tissue (Figure 3F).
Figure 3.
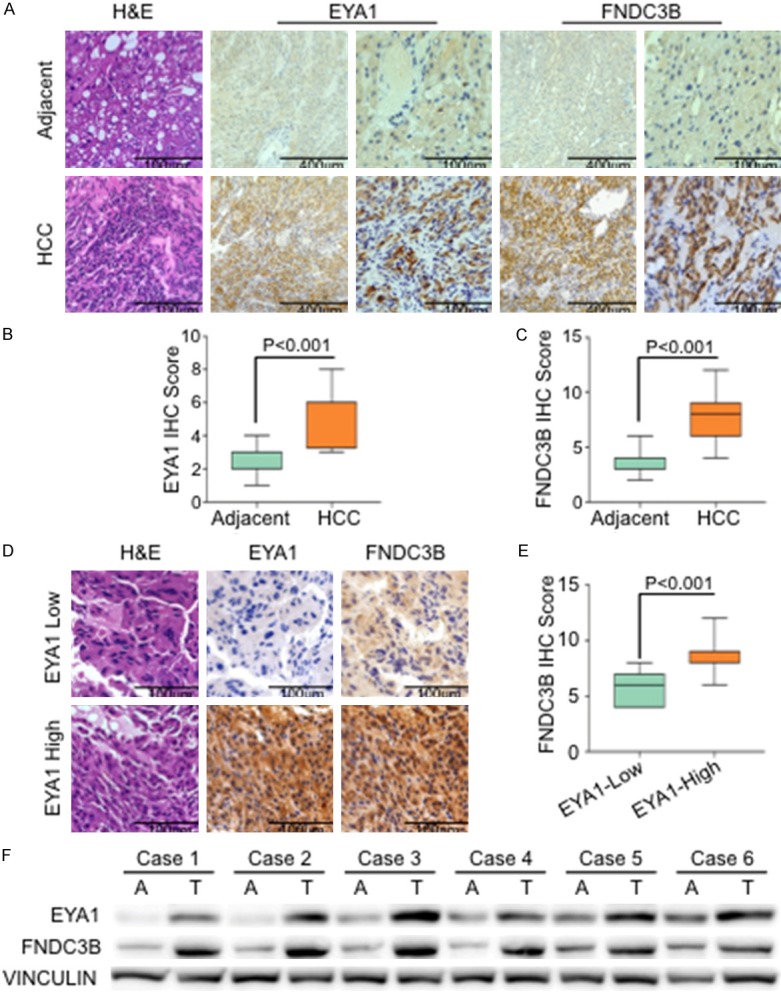
EYA1 was significantly associated with FNDC3B in human HCC. A: Representative images of the Immunohistochemistry (IHC) staining showed the expression of EYA1 and FNDC3B in HCC and adjacent normal tissue. B and C: Semi-quantitative analysis of EYA1 and FNDC3B in normal and HCC tissue. D: Representative IHC staining of EYA1 and FNDC3B in low-EYA1 (IHC score < 6) and high-EYA1 (IHC score ≥ 6), respectively. E: Semi-quantitative IHC scores of FNDC3B in low-EYA1 and high-EYA1 group. F: Western blot showed the expressions of EYA1 and FNDC3B in 6 paired of tumor (T) and adjacent tissue (A).
EYA1 contributed to the EMT in HCC
To detect the baseline expression of EYA1 in HCC cell, 4 different cell lines were examined by western blot analysis. After normalized by GAPDH, we have found that EYA1 was highly enriched in HepG2 and Huh-7 cells, whereas kept lower expression in Hep3B and SMMC-7721 cells (Figure 4A). In this study, we used HepG2 and Hep3B cell lines to generate EYA1 knock-in/knock-down in vitro model. We employed lentivirus-mediated gene transfer system to establish EYA1 overexpressing Hep3B cell lines by transduced pcDNA3.1-EYA1 (referred to as Hep3B-EYA1) and controls (referred to as Hep3B-Vector) plasmid. Meanwhile, non-targeting negative control or siRNA was transduced into HepG2 cell to silence EYA1 expression. qPCR and western blot analysis were used to confirm EYA1 knockdown efficiency in HepG2-siEYA1 cells and the overexpression of EYA1 in Hep3B-EYA1 cells (Figure 4B and 4C). In accordance with the previous findings, FNDC3B was identified as a downstream target of EYA1. Overexpression of EYA1 in Hep3B cells significantly increased the abundance of FNDC3B compared to the control group. Conversely, knockdown EYA1 in HepG2 cells could induce a reduction of FNDC3B (Figure 4B). To further evaluate the effect of EYA1 on EMT process, the epithelial markers including N-cadherin and E-cadherin were examined by Western Blot and Immunofluorescent (IF) staining. Transfecting the exogenous EYA1 gene into Hep3B cells can increase N-cadherin expression, and EYA1 knockdown resulted in a reduction of N-cadherin (Figure 4C). The data either from Western Blot or IF staining suggested that EYA1 significantly decreased the expression of E-cadherin (Figure 4C-E). These data indicated that EYA1 could cooperate with FNDC3B to induce the EMT-like phenotype change in HCC via increasing N-cadherin and decreasing E-cadherin.
Figure 4.
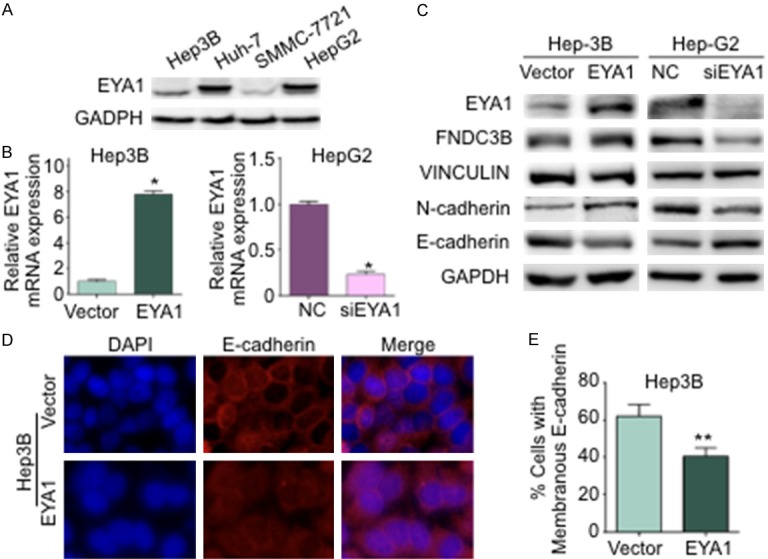
EYA1 contributed to the EMT in HCC. A: Western Blot showed the baseline expression of EYA1 in HCC cell lines, including Hep3B, Huh-7, SMMC-7721 and HepG2 cells. B: qPCR analysis confirmed the stable overexpression of EYA1 in Hep3B cells and the EYA1 knockdown efficiency in HepG2 cells. C: Western blot examined the expressions of EYA1, its downstream target FNDC3B, the EMT marker N-cadherin and E-cadherin. Vinculin and GAPDH were used as internal control. D: The representative image of immunofluorescence (IF) staining for E-Cadherin (red) in Hep3B-Vect or Hep3B-EYA1 cells. DAPI marked the nucleus. E: Quantification of the membranous E-Cadherin. *P < 0.05, **P < 0.01.
EYA1 increased cell migration and invasion in HCC
To identify the functional role of EYA1 in HCC, the wound healing and Transwell invasion assay were performed to investigate the migration and invasion ability. Wound healing assays indicated the Hep3B cell with ectopic expression of EYA1 showed faster closure (Figure 5A and 5B), whereas there was a significant reduction in migration of HepG2 cells after knockdown EYA1 by siRNA (Figure 5C and 5D). Similarly, the number of invaded cells was significantly increased after ectopic expression of EYA1 in comparison with the vector group (Figure 5E and 5F). Silencing EYA1 endogenous expression led to a noticeable reduction of invasion of HepG2 cells (Figure 5G and 5H). These results suggest that EYA1 is required for the cell migration and invasion of HCC.
Figure 5.
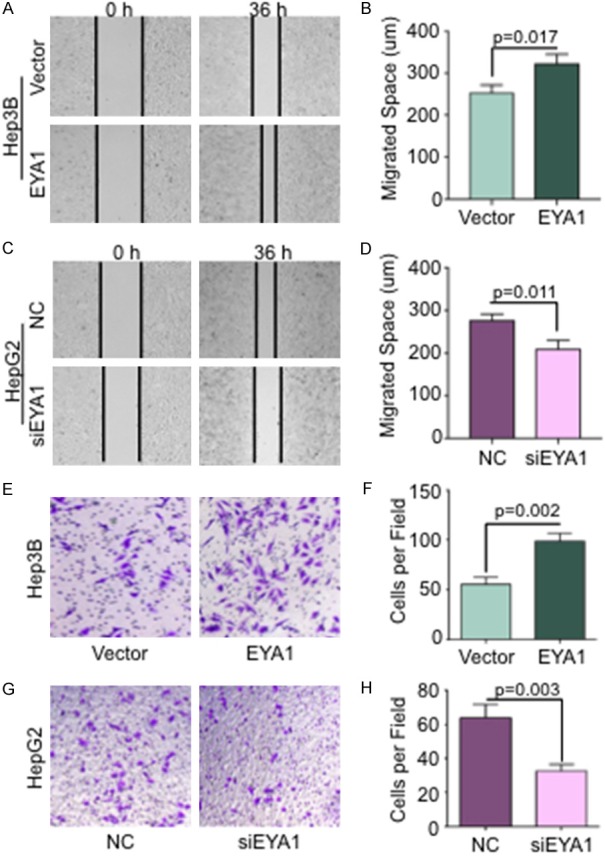
EYA1 increased cell migration and invasion in HCC cells. A: The representative images of wound-healing assay in Hep3B cells with EYA1 overexpression vs. vector group at 0 h and 36 h, respectively. B: Quantification of migrated distance in Hep3B cells. C: The representative images of wound-healing assay in HepG2 cells with EYA1 knockdown by siRNA compared to negative control (NC) at 0 h and 36 h, respectively. D: Quantification of migrated distance in HepG2 cells. E: The representative images of Transwell assay in Hep3B vector and EYA1 overexpression group. F: Quantification of Transwell invasion assay in Hep3B cells. G: The representative images of Transwell assay in HepG2 cell transfected by NC and EYA1 siRNA. H: Quantification of Transwell invasion assay in HepG2 cells.
Discussion
Hepatocellular carcinoma can be derived by the aberrant expressions of those genes and signal pathways related to growth and development. In this respect, EYA family acts as a critical transcription activator in Dach-Six-Eya axis and has been well addressed in eye formation of Drosophila. The overexpression of EYAs has been found in various cancer and perform a critical role in tumor growth and metastasis [14]. Our work took the initiative to demonstrate the role of EYA1 in HCC. In this study, we showed that EYA1 and FNDC3B were upregulated in HCC samples and high EYA1/FNDC3B was related to short survival time by analyzing multiple public microarray datasets as well as our cohort. In vitro, FNDC3B was identified as a novel downstream target of EYA1. EYA1 promoted cell migration and invasion via regulation EMT process in HCC.
Compared with the other two families of RDGN (SIX and DACH), the molecular regulation of EYAs is more complicated, since its dual roles as either protein phosphatase or transcriptional cofactor for SIX family. For example, recent evidence has been highlighted that Eya3 can control the stability of c-Myc via PP2A-B56α-mediated dephosphorylation on phospho-T58 (pT58) in breast cancer [28]. EYA1 also stabilized c-Myc protein abundance via decreasing the pT58 levels and mediated c-Myc ubiquitination via FBW-dependent manner [21]. Meanwhile, EYA can interact with SIX1 to induce EMT and metastasis, whereas disruption in the binding domain of the helix structure in EYA-SIX1 complex can diminish this effect [8]. But beyond that, whether EYAs exhibit other possible regulation independent of the existing mechanisms trigger extensive interest. In this study, FNDC3B was identified as a novel downstream target of EYA1 in regulating EMT process during HCC metastasis. EYA1 integrated with FNDC3B to increase the migration and invasion in HCC cells, which may be dependent on the regulation of N-cadherin and E-cadherin expression. However, we didn’t exclude the possibilities that the tyrosine phosphatases or SIX1 are required for the regulation of FNDC3B by EYA1. Therefore, the next challenge is exploring the molecular basis underlying the connection between EYA1 and FNDC3B in HCC development.
FNDC3B also called FAD104 (factor for adipocyte differentiation 104), is involved in adipocyte and osteoblast differentiation, and lung maturation by regulating cell adhesion, growth, and migration [25,26]. To date, the research of FNDC3B in cancer is still in a preliminary stage, and the studies from different groups have produced conflicting results. FNDC3B was found to suppress the invasion and metastasis of melanoma cells by inhibition of STAT3 activity [29]. On the contrary, an integrated microarray study by overlapping the common genes with a changed expression in 20 different cancer types indicated that FNDC3B was one of the up-regulated genes in all cancerous tissue [30]. Another evidence supporting FNDC3B as an oncogene was the overexpression of FNDC3B were found in over or close to 50% of esophageal, lung squamous and ovarian cancers, which was tightly associated with EMT process and activation several key pathways in tumorigenesis and tumor progression, including PI3-kinase/Akt, Rb1 and TGFβ signaling [31]. In HCC development, only one group has reported that FNDC3B can promote tumor cells migration and invasion, but the molecular mechanism is still largely unknown [27]. One novel finding from our study was FNDC3B was identified as a poor predictor in the clinical outcome of HCC patients. Besides, FNDC3B performed as a downstream target of the oncogenic EYA1 in regulation the EMT process of HCC. Thus, it is convincible that FNDC3B is a critical mediator in EYA-dependent oncogenesis.
Although most of the studies support the role of EYA as a tumor promoter in breast and ovarian cancer [18,32], in pancreatic cancer, EYA2 showed the opposite effect, which decreased the tumor growth [33], suggesting EYAs has its specificity in relation to different cell or tissue. Another interesting finding from our study was EYA1 increased expression in HCC compared to normal liver tissue confirmed by the analysis from both multiple public microarray datasets and our cohorts. Our study further indicated EYA1 was an independent prognostic factor for HCC patients and a combined evaluation of EYA1 and FNDC3B profiles would provide more accurate survival prediction. Intriguingly, EYAs have recently become the novel molecular targets for chemotherapy. To antagonize the tyrosine phosphatases activity of EYA protein, seven novel classes of compounds were found through structure-based virtual screening technology [34]. A recent finding suggested that EYA1 integrated with hypoxia-inducible factor 1 (HIF-1α) to induce VEGF-A expression, resulting in tumor angiogenesis in the colorectal tumor [35]. In this respect, Benzbromarone and its derivate, Benzarone were found to be the EYA inhibitors [36], and 6-hydroxy Benzbromarone inhibited angiogenesis by blocking the activity of EYAs [37]. Earlier evidence has raised the possibility that targeting upstream signal of EYAs by small molecular inhibitors, like PI3K/Akt pathway, would bring potential clinical benefit to specific subpopulation [38]. Importantly, the activator of the Wnt signal pathway, lithium chloride was identified to reduce the expression of EYA1 and SIX2 [39]. EYAs are also involved in immune regulation during tumor development. Recently, Dr. Ford has found that Eya3 coordinated with PD-L1 to induce immune suppression via decreasing CD8+ T cell amount in triple-negative breast cancer [40]. Therefore, therapies targeting EYAs may have a broad effect and be a more effective and durable treatment to advanced HCC patients.
In conclusion, we demonstrate that EYA1 was upregulated in HCC tissues and closely correlated with the poor outcome of patients. More importantly, our research indicated that EYA1 contributed to the progression of HCC via activation of FNDC3B and induced EMT-related genes expression. Comprehensively evaluation of EYA1 and FNDC3B profiles could provide novel insight into the molecular mechanism underlying HCC as well as develop specific therapeutic strategies for personal treatment.
Disclosure of conflict of interest
None.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 4.Mishra L, Banker T, Murray J, Byers S, Thenappan A, He AR, Shetty K, Johnson L, Reddy EP. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu H, Zhang C, Yin T, Wu K. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int J Cancer. 2016;138:1067–1075. doi: 10.1002/ijc.29560. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Kong D, Wu H, Yuan X, Xu H, Zhang C, Wu G, Wu K. Interplay of retinal determination gene network with TGF-β signaling pathway in epithelial-mesenchymal transition. Stem Cell Investig. 2015;2:12. doi: 10.3978/j.issn.2306-9759.2015.05.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat Struct Mol Biol. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 10.Musharraf A, Kruspe D, Tomasch J, Besenbeck B, Englert C, Landgraf K. BOR-syndrome-associated Eya1 mutations lead to enhanced proteasomal degradation of Eya1 protein. PLoS One. 2014;9:e87407. doi: 10.1371/journal.pone.0087407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blevins MA, Towers CG, Patrick AN, Zhao R, Ford HL. The SIX1-EYA transcriptional complex as a therapeutic target in cancer. Expert Opin Ther Targets. 2015;19:213–225. doi: 10.1517/14728222.2014.978860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. The eyes absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong D, Liu Y, Liu Q, Han N, Zhang C, Pestell RG, Wu K, Wu G. The retinal determination gene network: from developmental regulator to cancer therapeutic target. Oncotarget. 2016;7:50755–50765. doi: 10.18632/oncotarget.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Ohgi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates six1-dach-eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 16.Yuan B, Cheng L, Chiang HC, Xu X, Han Y, Su H, Wang L, Zhang B, Lin J, Li X, Xie X, Wang T, Tekmal RR, Curiel TJ, Yuan ZM, Elledge R, Hu Y, Ye Q, Li R. A phosphotyrosine switch determines the antitumor activity of ERβ. The J Clin Invest. 2014;124:3378–3390. doi: 10.1172/JCI74085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, Friedman R, Klein U, Tycko B. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu K, Li Z, Cai S, Tian L, Chen K, Wang J, Hu J, Sun Y, Li X, Ertel A, Pestell RG. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013;73:4488–4499. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikpour P, Emadi-Baygi M, Emadi-Andani E, Rahmati S. EYA1 expression in gastric carcinoma and its association with clinicopathological characteristics: a pilot study. Med Oncol. 2014;31:955. doi: 10.1007/s12032-014-0955-y. [DOI] [PubMed] [Google Scholar]
- 20.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, Ji H, Thirman MJ. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Rodriguez Y, Cheng C, Zeng L, Wong EY, Xu CY, Zhou MM, Xu PX. EYA1’s conformation specificity in dephosphorylating phosphothreonine in Myc and its activity on Myc stabilization in breast cancer. Mol Cell Biol. 2017;37:e00499–16. doi: 10.1128/MCB.00499-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Kong D, Liu Y, Cui Q, Wang K, Zhang D, Wang J, Zhai M, Yan J, Zhang C, Wu G. Sapylin promotes wound healing in mouse skin flaps. Am J Transl Res. 2017;9:3017–3026. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu H, Guo M, Yu S, Zhang C, Yin T, Wu K. DACH1 is a novel predictive and prognostic biomarker in hepatocellular carcinoma as a negative regulator of Wnt/β-catenin signaling. Oncotarget. 2015;6:8621–8634. doi: 10.18632/oncotarget.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 25.Obara M, Sakuma T, Fujikawa K. The third type III module of human fibronectin mediates cell adhesion and migration. J Biochem. 2010;147:327–335. doi: 10.1093/jb/mvp168. [DOI] [PubMed] [Google Scholar]
- 26.Nishizuka M, Kishimoto K, Kato A, Ikawa M, Okabe M, Sato R, Niida H, Nakanishi M, Osada S, Imagawa M. Disruption of the novel gene fad104 causes rapid postnatal death and attenuation of cell proliferation, adhesion, spreading and migration. Exp Cell Res. 2009;315:809–819. doi: 10.1016/j.yexcr.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Lin CH, Lin YW, Chen YC, Liao CC, Jou YS, Hsu MT, Chen CF. FNDC3B promotes cell migration and tumor metastasis in hepatocellular carcinoma. Oncotarget. 2016;7:49498–49508. doi: 10.18632/oncotarget.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Zhou H, Li X, Vartuli RL, Rowse M, Xing Y, Rudra P, Ghosh D, Zhao R, Ford HL. Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat Commun. 2018;9:3830. doi: 10.1038/s41467-018-06265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh D, Nishizuka M, Osada S, Imagawa M. Fad104, a positive regulator of adipocyte differentiation, suppresses invasion and metastasis of melanoma cells by inhibition of STAT3 activity. PLoS One. 2015;10:e0117197. doi: 10.1371/journal.pone.0117197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Yi Y, Liu P, Wen W, James M, Wang D, You M. Common human cancer genes discovered by integrated gene-expression analysis. PLoS One. 2007;2:e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai C, Rajaram M, Zhou X, Liu Q, Marchica J, Li J, Powers RS. Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle. 2012;11:1773–1781. doi: 10.4161/cc.20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, Barchetti A, Vezzani C, O’Brien-Jenkins A, Wang J, Ward MR, Courreges MC, Fracchioli S, Medina A, Katsaros D, Weber BL, Coukos G. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. 2005;65:925–932. [PubMed] [Google Scholar]
- 33.Vincent A, Hong SM, Hu C, Omura N, Young A, Kim H, Yu J, Knight S, Ayars M, Griffith M, Van Seuningen I, Maitra A, Goggins M. Epigenetic silencing of EYA2 in pancreatic adenocarcinomas promotes tumor growth. Oncotarget. 2014;5:2575–2587. doi: 10.18632/oncotarget.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H, Jung SK, Yu KR, Kim JH, Kim YS, Ko JH, Park BC, Kim SJ. Structure-based virtual screening approach to the discovery of novel inhibitors of eyes absent 2 phosphatase with various metal chelating moieties. Chem Biol Drug Des. 2011;78:642–650. doi: 10.1111/j.1747-0285.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- 35.Cai S, Cheng X, Liu Y, Lin Z, Zeng W, Yang C, Liu L, Chukwuebuka OA, Li W. EYA1 promotes tumor angiogenesis by activating the PI3K pathway in colorectal cancer. Exp Cell Res. 2018;367:37–46. doi: 10.1016/j.yexcr.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 36.Tadjuidje E, Wang TS, Pandey RN, Sumanas S, Lang RA, Hegde RS. The EYA tyrosine phosphatase activity is pro-angiogenic and is inhibited by benzbromarone. PLoS One. 2012;7:e34806. doi: 10.1371/journal.pone.0034806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey RN, Wang TS, Tadjuidje E, McDonald MG, Rettie AE, Hegde RS. Structure-activity relationships of benzbromarone metabolites and derivatives as EYA inhibitory anti-angiogenic agents. PLoS One. 2013;8:e84582. doi: 10.1371/journal.pone.0084582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Kaneko S, Li XK, Li X. The PI3K/Akt signal hyperactivates Eya1 via the SUMOylation pathway. Oncogene. 2015;34:2527–2537. doi: 10.1038/onc.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price KL, Kolatsi-Joannou M, Mari C, Long DA, Winyard PJD. Lithium induces mesenchymal-epithelial differentiation during human kidney development by activation of the Wnt signalling system. Cell Death Discov. 2018;4:13. doi: 10.1038/s41420-017-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vartuli RL, Zhou H, Zhang L, Powers R, Klarquist J, Rudra P, Vincent MY, Ghosh D, Costello JC, Kedl RM, Slansky JE, Zhao R, Ford HL. Eya3 promotes breast tumor-associated immune suppression via threonine phosphatase-mediated PD-L1 upregulation. J Clin Invest. 2018;128:2535–2550. doi: 10.1172/JCI96784. [DOI] [PMC free article] [PubMed] [Google Scholar]


