Figure 3.
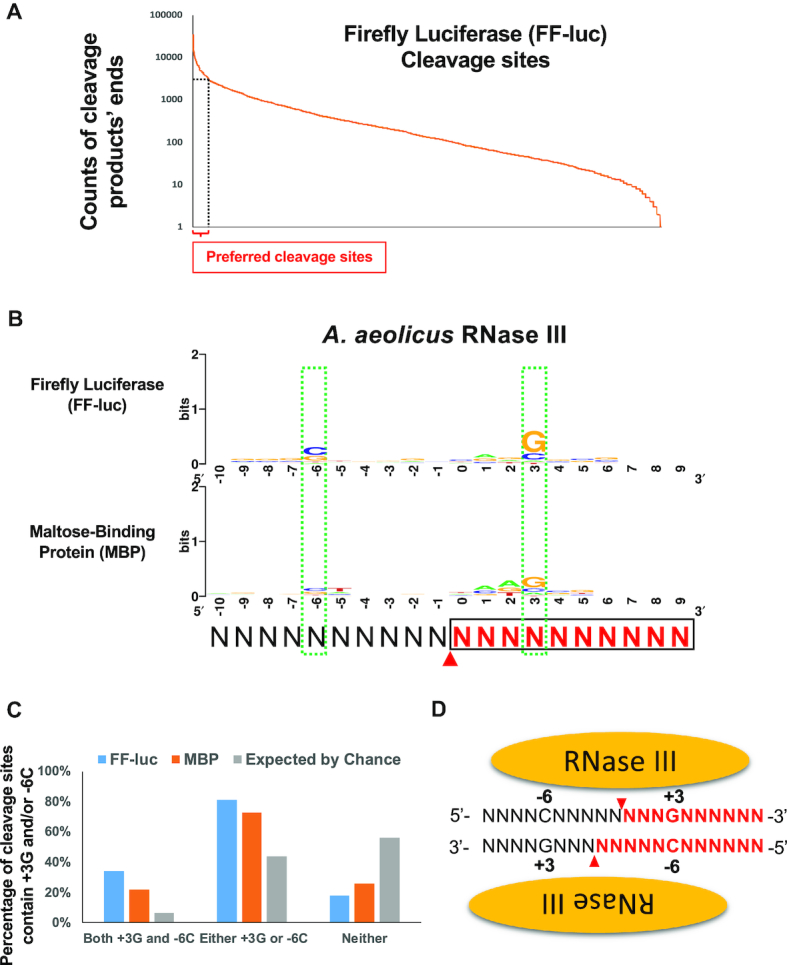
RNase III recognizes +3G in cleavage site selection. (A) Cleavage sites were inferred by the ends of cleavage products. Cleavage sites along FF-luc substrate were sorted by the counts of cleavage products’ ends and plotted. Cleavage sites supported by a count higher than 2,907, which is 5× the average (581.3), were defined as preferred cleavage sites (hotspots) and indicated by the dashed lines. (B) Sequence logos surrounding preferred AaRNase III cleavage sites on FF-luc and MBP are presented. Red arrowheads indicate the cleavage site (between positions 0 and -1), which is inferred from the end of cleavage products. Cleavage product upstream of cleavage site is in black, whereas cleavage products downstream of cleavage site are in red and boxed. Two green dashed-line boxes indicate two consensus nucleotides +3G and -6C. (C) Percentage of cleavage sites that contain +3G and/or -6C is higher than that expected by chance. (D) Schematic illustration of the RNase III:dsRNA complex. The +3G and -6C are symmetric relative to the cleavage sites that are indicated by red arrowheads. Cleavage products on one side of cleavage site are in black, whereas cleavage products on the other side of cleavage site is in red.
