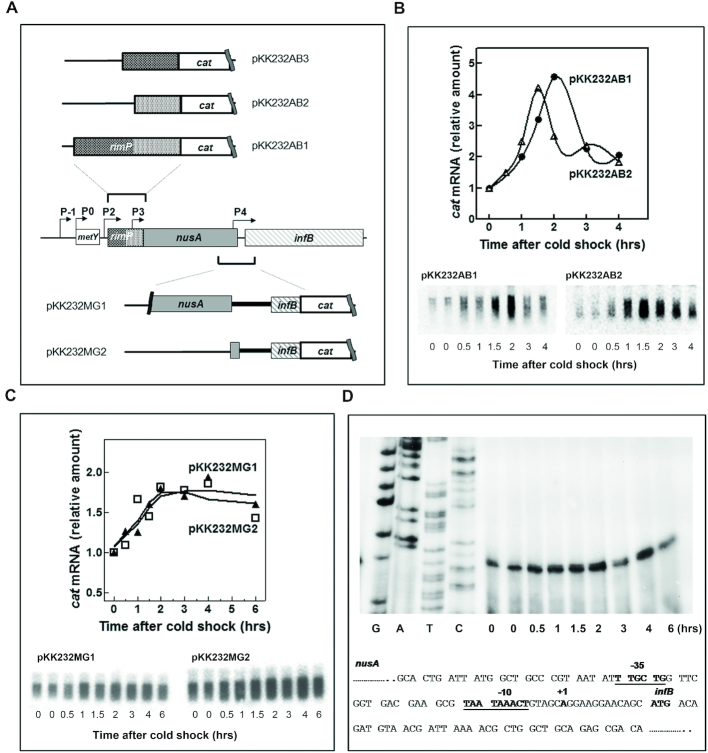
Figure 7.
Identification of P3 and P4 two new cold-inducible promoters in the nusA-infB operon. (A) Representation of the cat transcriptional fusion constructs used for the identification of new promoters. The chromosomal DNA fragments used in the transcriptional fusions with the cat gene are indicated in the panel. The fragments corresponding to the entire rimP and to the proximal and distal regions of the same gene and those corresponding to the nusA-infB intragenic regions are schematically indicated above and below the nusA-infB operon, respectively. (B) Steady state levels of cat mRNA present before and after the indicated times of cold stress in cells transformed with pKK232AB1(•) and pKK232AB2 (△). The RNA levels were estimated from the quantification of the hybridization bands detected by northern blotting like those shown below the graph. (C) Cold stress-dependent increase of the steady state levels of cat mRNA in cells carrying plasmids in which the promoter-less cat gene is placed downstream the proximal portion of infB plus a long (pKK232MG1, ▴) or a short (pKK232MG2, □) DNA segment belonging to the distal region of nusA. (D) Primer extension analysis of the start point of the transcript originating from P4, the promoter located in the nusA-infB intragenic region whose sequence is reported below. The identified core elements of the P4 promoter and the first transcribed base are indicated in bold letters. The curves shown in panels (B) and (C) have been fitted using the fitspline/lowess program (Graphpad Prism).