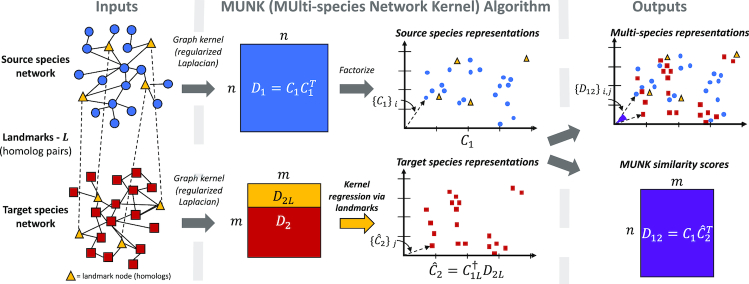
Figure 1.
Given a source PPI network, a target PPI network, and a set of landmark (homolog) pairs across species, MUNK computes diffusion kernels for each network. Then, MUNK factorizes the diffusion kernel for the source species into its reproducing kernel Hilbert space (RKHS). Finally, MUNK solves a linear system of the source species’ RKHS and the target species’ diffusion kernel to create a multi-species vector embedding of source and target proteins. The inner products of these embeddings correlate with functional similarities and the embeddings themselves allow for functional comparisons between proteins across the two networks.