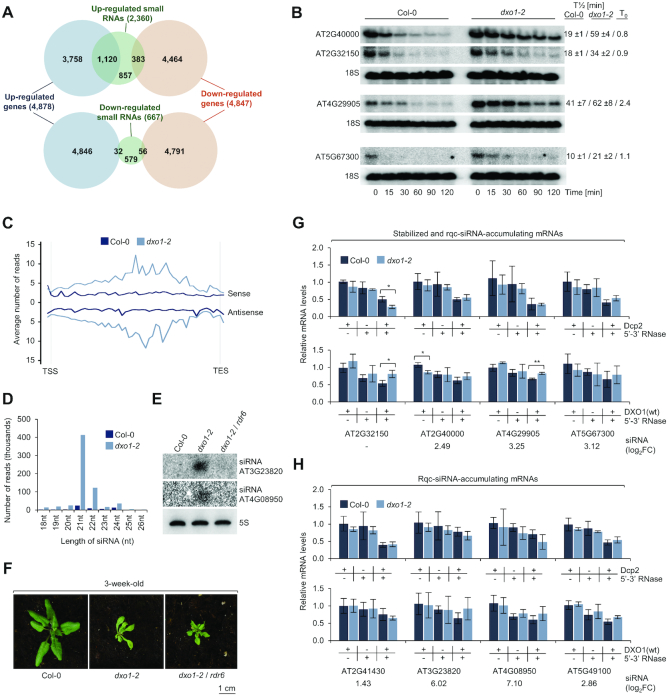
Figure 5.
RNA quality control siRNAs (rqc-siRNAs) accumulate in dxo1 plants. (A) Overlap between genes with affected expression (FDR < 0.05, |log2FC| > 0) and genes with affected sRNA levels in dxo1-2 (FDR < 0.05, |log2FC| > 1). Numbers represent genes identified in RNA-seq and sRNA-seq experiments and overlapping genes. (B) Half-lives of selected mRNAs by northern blot after transcriptional inhibition by cordycepin. T1/2, mean values of transcript levels relative to untreated plants; T0, relative level of transcript in dxo1-2 versus Col-0 at zero time point; based on three biological replicates. 18S rRNA; loading control. (C) Small RNAs in dxo1-2 originate from both mRNA strands. Average read counts over protein-coding nuclear genes with siRNA up-regulation (FDR < 0.05, |log2FC| > 1, n = 2,288 loci). (D) Up-regulated siRNAs divided according to length. (E) Small RNA detection by northern blotting; 5S rRNA, loading control. (F) Three-week-old Col-0, dxo1-2 and dxo1-2/rdr6 plants. (G, H) Stabilized (G) and rqc-siRNA-enriched (H) genes do not show defects in mRNA deNADding and capping. Total RNA treated with indicated combinations of DXO1, Dcp2 and Terminator was analyzed by RT-qPCR. mRNA levels were normalized to Pol III-transcribed pre-tRNA-snoRNA (tsnoRNA) negative control and calculated relative to untreated Col-0 and dxo1-2. Results are the mean of three biological replicates, error bars represent standard deviation; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (t-test); siRNA accumulation from sRNA-seq (log2FC) is indicated below the graph. See Supplementary Figure S5 for additional information.