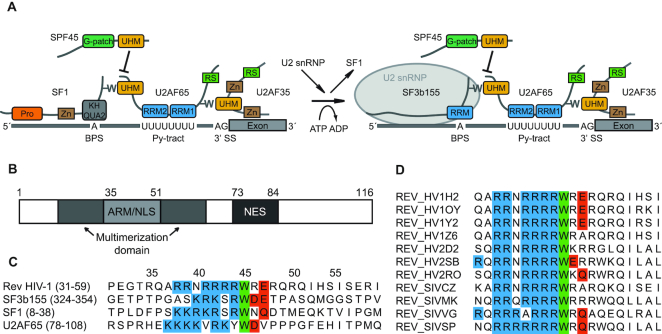
Figure 1.
HIV-1 Rev protein has a conserved ULM resembling motif. (A). Schematic representation of ULM–UHM interactions required for constitutive and alternative splicing. Pro – proline-rich region; Zn – zinc-finger domain; BPS – branch point sequence; Py – polypyrimidine; RS – arginine–serine-rich domain. (B). Domain organization of full-length Rev. (C). Alignment of ULM sequences of HIV-1 Rev, SF3b155 (ULM5), SF1 and U2AF65. Conserved residues are marked: blue – basic residues preceding the conserved tryptophan; green – conserved tryptophan; red – acidic and N/Q-type residues following the conserved tryptophan. (D). Alignment of the ULM motif in selected HIV-1, HIV-2 and SIV strains. With the exception of single HIV-2 and SIV strains, the tryptophan is strictly conserved in all aligned isolates (a total of 2000 HIV-1, HIV-2 and SIV sequences was used for the alignment).