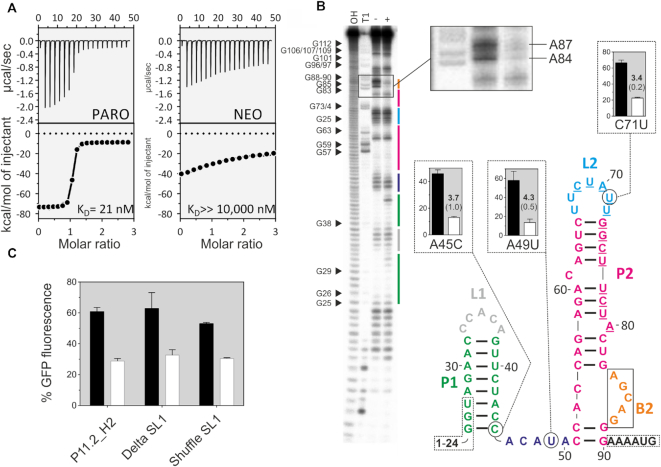
Figure 4.
Biochemical characterization of the PARO riboswitch. (A) Determination of the binding constant of P11.2H2 with ITC against PARO (left) and NEO (right). Top panels: power required to maintain the temperature of the RNA solution recorded over the time until saturation was reached (baseline-corrected). Bottom panel: integrated heats of interaction plotted against the molar ratio of ligand over RNA and fitted to a single binding site model (MicroCal PEAQ-ITC Analysis Software 1.1.0). (B) In-line probing experiment for PARO-RS. Shown is the cleavage pattern in the absence (−) and presence (+) of 10 μM PARO under alkaline conditions. As references and for nucleotide position assignment, hydroxyl reaction (OH) and nuclease T1 digestion (T1) were loaded onto the gel. G nucleotides are highlighted. Colour coding for identified stem and loop regions follows the coding for the proposed secondary structure. The docking sequence is represented by the underlined nucleotides (68–80). 5′ and 3′ primer binding site regions are framed with dotted lines. Proposed secondary structure of the PARO-RS riboswitch including two stems (P1 and P2), two loop regions (L1 and L2), an asymmetric internal bulge (B2) and an unstructured spacer region (46–50). The mutated nucleotides compared to P11.2H2 are encircled. (C) Comparison of the GFP gene regulation between the P11.2H2 candidate and the constructs Delta S1 and Shuffle S1. As observed, none of the constructions delete the switching properties, but slightly reduce the switching factor. This indicates that the S1 plays probably a minor role in the switching properties of the riboswitch.