Figure 12.
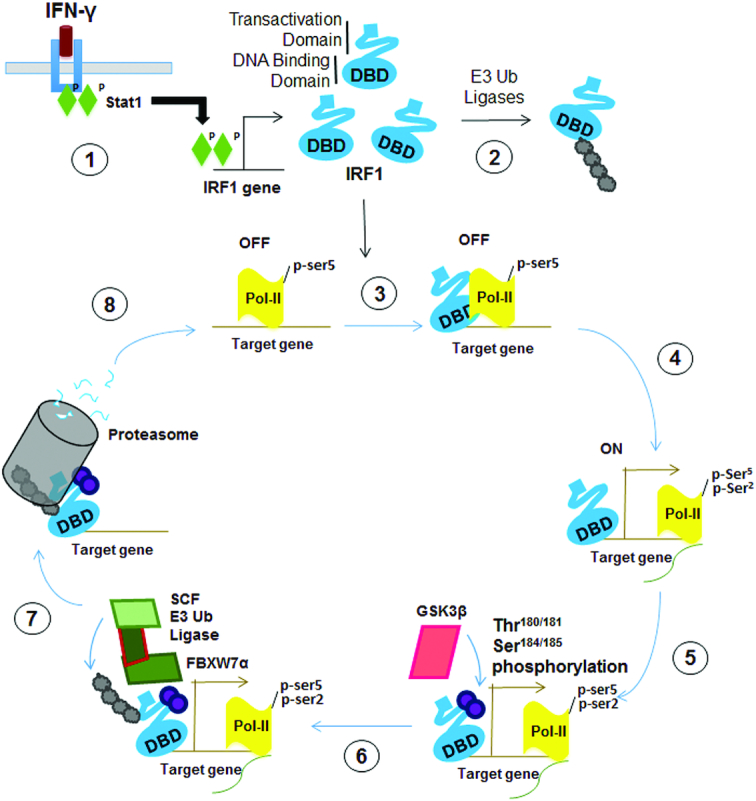
Schematic depicting a proposed model of the regulation of IRF1 activity by GSK3 kinases. 1) External stimuli (such as IFNγ signalling activate STAT1 leading to increase expression of cellular IRF1 protein. 2) Steady state levels of IRF1 protein are maintained by the Ub proteasome system. Non-DNA bound IRF1 is ubiquitinated at lysine residues exposed within the DBD. 3) Nuclear IRF1 binds recognition sequences in target promoters. In many cases such as TBC1D32 gene, these IRF1-bound promoters are marked by high levels of pSer5 (initiating) modified RNA-Pol-II and are thus poised for transcription. Engagement with DNA shields the lysines within the DBD from recognition by E3 ligases and subsequent degradation by the proteasome. This allows time for further events that are necessary for transcription to occur. 4) Transcription is initiated, RNA-Pol-II is marked with pSer2 (elongating). 5) Successful initiation triggers phosphorylation of IRF1 at T181/S185 by GSK3β. It is not known how this phosphorylation senses RNA-Poll firing, perhaps a reorganization of proteins at the promoter unmasks epitopes in IRF1 allowing binding and phosphorylation. 6) Phosphorylation of IRF1 generates a phospho-degron recognized by SCFFbxw7α, which promotes K48 linked ubiquitination of IRF1. 7) IRF1 is degraded by the proteasome. It is not known if the degradation occurs while IRF1 is bound to DNA, or if a release of IRF1 occurs beforehand. 8) The previously occupied element is now free for additional molecules of IRF1 (or other proteins) to re-bind and begin a new cycle of transcription. Local concentrations of IRF1 protein – determined by the balance between de novo generation of IRF1 and degradation will help dictate whether this additional cycle occurs.
