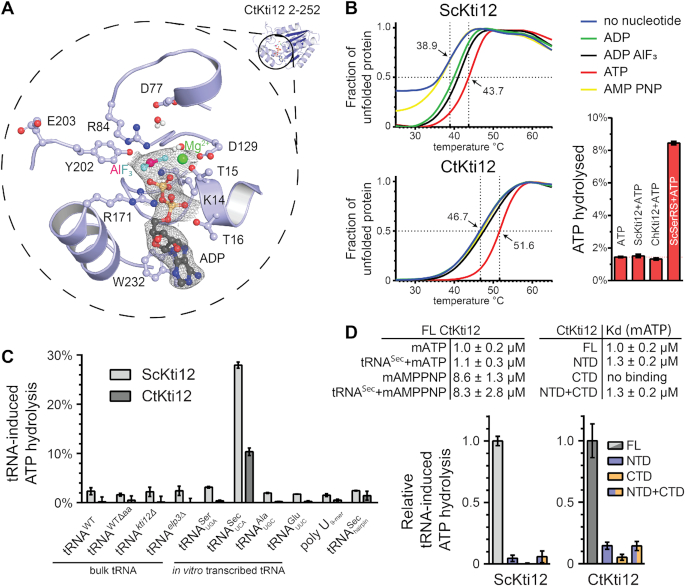
Figure 2.
Structural and functional characterization of ATP hydrolysis by Kti12. (A) A structural close-up of the ATP-binding pocket with important residues shown in ball-and-stick representation. Isomesh represents an OMIT map at σ = 1.0 around the ligand. Aluminum fluoride (magenta and cyan) and magnesium (green) are shown and the residues involved are labeled. (B) Thermal shift assays with ScKti12 (top) and CtKti12 (bottom) in the presence/absence of nucleotides. ATP hydrolysis (bottom right) was investigated using the malachite green method (right). Yeast seryl-tRNA synthetase was used as a positive control for basal ATP hydrolysis relative to ATP alone. (C) ATP hydrolysis of ScKti12 and CtKti12 in response to bulk or in vitro transcribed tRNAs (see labeling). All in vitro transcribed tRNAs possess 3′ CCA. Deamino-acylated (Δaa) tRNA was obtained by deamino-acylation of bulk tRNA obtained from WT yeast cells. A poly U (nine bases) and a tRNASec anticodon arm hairpin were used as a specificity control. Reactions contained tRNA and protein mixed at 1:1 stoichiometry and were incubated for 15 hours at 37°C. Results are presented as the percent of ATP hydrolyzed and calculated standard deviations are shown (n = 3). (D) Similarly, to (C), tRNASec was used to induce ATP hydrolysis in FL proteins, NTDs, CTDs or 1:1 stoichiometric mixture of NTDs and CTDs and calculated standard deviations are shown (n = 3).