Figure 7.
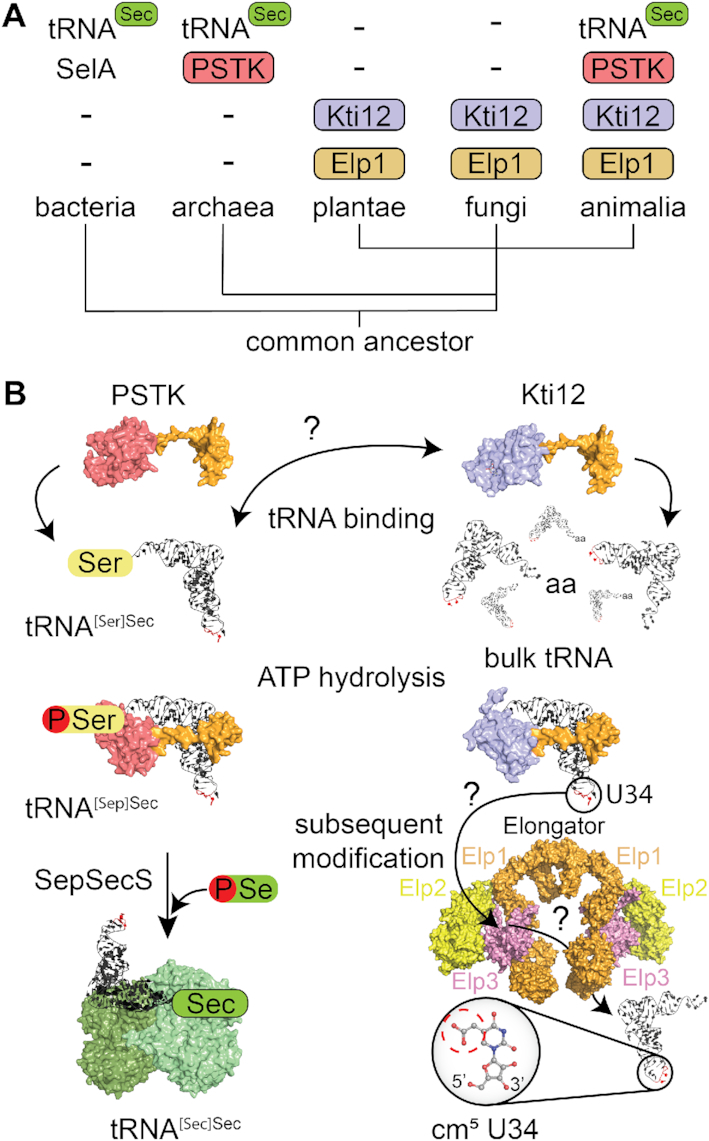
A comparison of Kti12 and PSTK analogs. (A) Schematic overview of the phylogenetic analyses of PSTK and Kti12 distribution. With the exception of bacteria, which use SelA protein for selenocysteine synthesis, PSTK appears together with a tRNASec in archaea and animalia. Plantae and fungi are missing both PSTK as well as tRNASec. Kti12 can be found within all eukaryotic lineages, which is also true for its interaction partner Elp1. (B) PSTK and Kti12 architectures are very similar. Both proteins bind tRNA, however, unlike PSTK, Kti12 is capable of binding to bulk yeast tRNA. Both PSTK and Kti12 utilize tRNA binding for induction of ATP hydrolysis. PSTK transfers gamma phosphate from ATP to tRNA[Ser]Sec generating tRNA[Sep]Sec, which is further used as a substrate by SepSecS, a tetrameric enzyme capable of using selenophosphate as a selenium donor for formation of Sec-tRNASec. Two catalytically active subunits of human SepSecS are shown in light green while catalytically inactive subunits are dark green. In case of Kti12, ATP hydrolysis enables cm5U34 modification in the anticodon of 11 tRNA species by the Elongator complex.
