Figure 3.
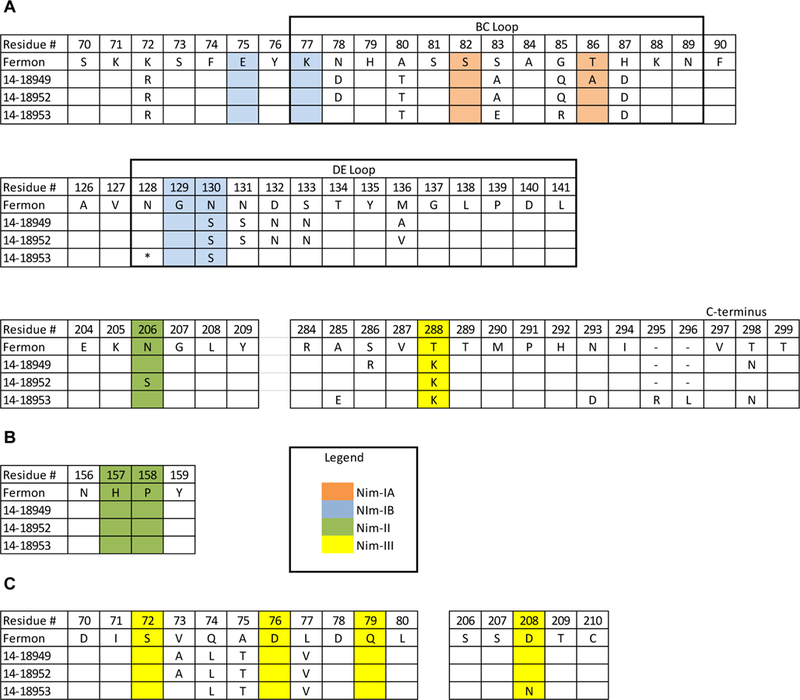
Protein sequence alignment of putative antigenic sites for EV-D68 viruses used in microneutralization assay. Alignments of EV-D68 capsid proteins VP1 (A), VP2 (B), and VP3 (C); only amino acid residues that differ from Fermon virus are shown. Residue number for each amino acid chain is based on the predicted cleavage sites from Liu et al [8]. Putative antigenic sites based on RV-14 are shaded as follows: NIm-IA (orange), Nim-IB (blue), Nim-II (green), and Nim-III (yellow).
* denotes an amino acid deletion at residue 140 in 14–18953.
- denotes a gap in the sequence alignment due to a two-amino-acid insertion in 14–18953.
