Fig. 3. Active site structure in PETG-liganded E. coli β-galactosidase.
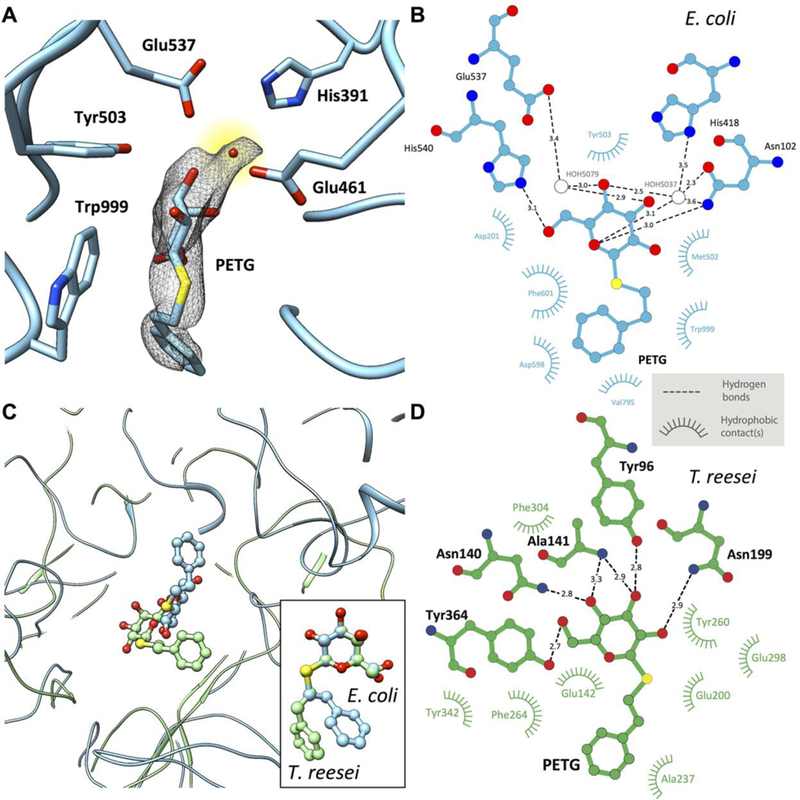
(A) Uncorrected cryo-EM density map showing density for PETG, an associated water molecule and five of the amino acids that line the binding pocket. The density for the PETG ligand appears to be at lower resolution than the surrounding amino acids, presumably because of some wobble in the binding pocket. (B) Plot of distances of various parts of PETG to residues in the vicinity of (β-galactosidase from E. coli determined using LIGPLOT (www.ebi.ac.uk/thornton-srv/software/LIGPLOT/). (C) Superposition of the ligand binding pocket structures in (β-galactosidase from E. coli (cyan, determined by cryo-EM at 2.2 Å resolution) and T. reesei (green, determined by x-ray crystallography at 1.4 Å resolution) illustrating the differences in protein and ligand structures, and comparison between the corresponding configurations of PETG (inset). (D) Plot of distances of various parts of PETG to amino acids in the vicinity of β-galactosidase from T. reesei determined using LIGPLOT.
