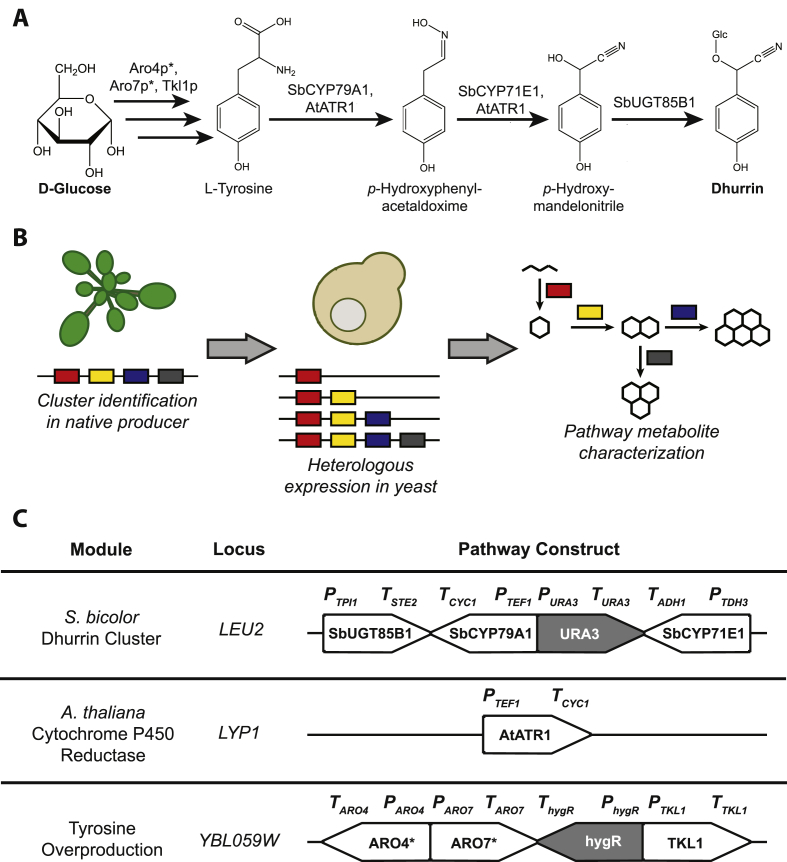
Fig. 1.
A heterologous metabolic pathway for dhurrin biosynthesis in yeast. A: The proposed dhurrin biosynthetic pathway. B: Rationale for heterologous pathway reconstruction. Biosynthetic gene clusters identified in plant genomes can be rapidly reconstructed in model microbes such as yeast, enabling rapid characterization of cluster enzymes in a clean metabolite background. C: Modular strategy for pathway construction. Genes in gray are selection markers. Abbreviations: Glc, glucose; URA3, S. cerevisiae orotidine-5′-phosphate decarboxylase; AtATR1, Arabidopsis thaliana cytochrome P450 reductase; ARO4*, S. cerevisiae 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate synthase Q166K; ARO7*, S. cerevisiae chorismate mutase T226I; TKL1, S. cerevisiae transketolase; hygR, hygromycin B phosphotransferase.