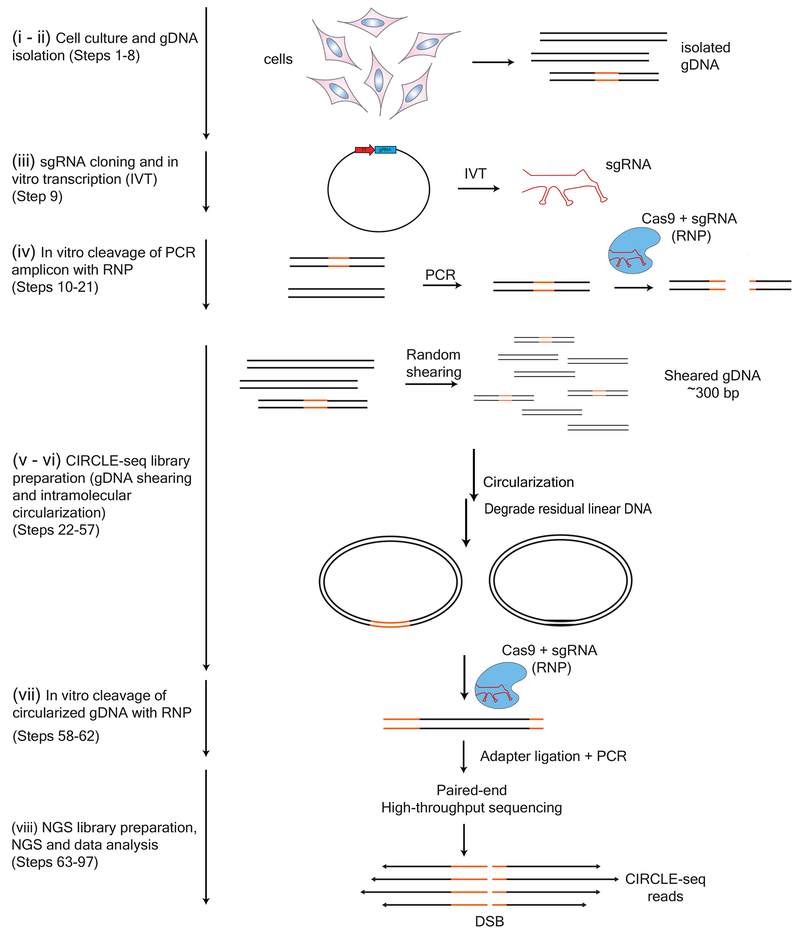
Figure 1 |.
Overview of CIRCLE-seq workflow. (i-ii) Genomic DNA is isolated from cells (Steps 1-8). (iii) sgRNA is cloned into an in vitro transcription plasmid and in vitro transcribed (Step 9). (iv) Cas9:sgRNA RNP complex is first functionally tested for its ability to functionally cleave a PCR amplicon containing the intended target site to near completion (Steps 10-21). (v-vi) gDNA is sheared in an average length of 300 bp and circularized by intramolecular ligation in a procedure outlined in detail in Figure 2. Remaining uncircularized (linear) DNA molecules are degraded with exonuclease treatment (Steps 22-57). (vii) Circular DNA molecules containing on- and off-target sites can subsequently be linearized with Cas9, releasing newly cleaved DNA ends (Steps 58-62) for (viii) adapter ligation, PCR amplification, and paired-end high-throughput sequencing (Steps 63-97). (Figure adapted with permission from Tsai et al, 20179.)