Figure 1.
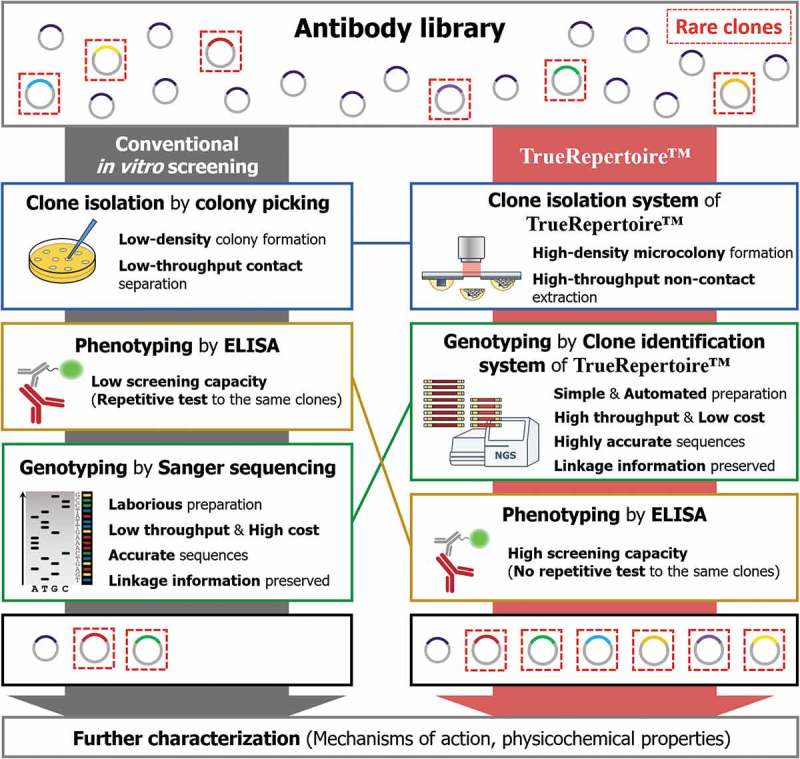
Comparison of the workflow between conventional in vitro screening and TrueRepertoire™. In the conventional in vitro screening process, clones in an antibody library are isolated by the colony picking method. The isolated clones are tested for their ability to bind to a target antigen by ELISA. Positive clones confirmed to have a binding activity are selected; then, the genotype of the clones is identified through Sanger sequencing. The low throughput and labor intensiveness of colony picking and Sanger sequencing limit the throughput. Moreover, the strategy of performing genotyping after phenotyping provokes the implementation of repetitive ELISA tests on the same clones, which reduces screening capacity. Overall, through the conventional in vitro screening process, in many cases, only a handful of clones are identified and retrieved for the following characterization. TrueRepertoire™ replaces colony picking with a new clone isolation system that enables the formation of microscale colonies in high density and extraction of the microcolonies in high throughput, without contact. The isolated clones are analyzed by the clone identification system of TrueRepertoire™. Through the clone identification system, highly accurate sequences of the clones are obtained based on the linkage information between heavy and light chains. Then, clones are selected based on the sequence information and tested for their binding activity by ELISA. Through the process, many clones, including rare clones, in the library can be identified and retrieved for further characterization.
