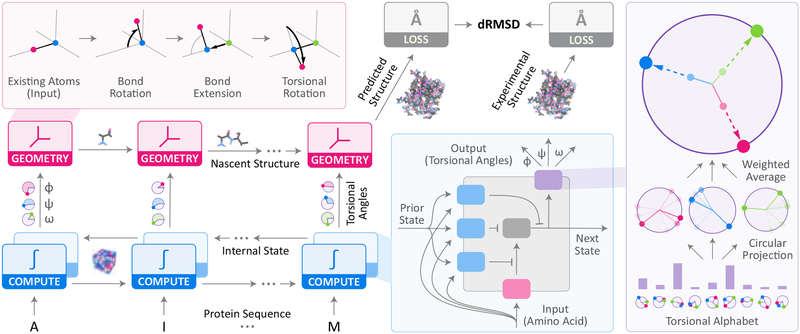
Figure 2: Recurrent geometric networks.
Protein sequences are fed one residue at a time to the computational units of an RGN (bottom-left), which compute an internal state that is integrated with the states of adjacent units. Based on these computations, torsional angles are predicted and fed to geometric units, which sequentially translate them into Cartesian coordinates to generate the predicted structure. dRMSD is used to measure deviation from experimental structures, serving as the signal for optimizing RGN parameters. Top-Left Inset: Geometric units take new torsional angles and a partial backbone chain, and extend it by one residue. Bottom-Right Inset: Computational units, based on Long Short-Term Memory (LSTMs) (Hochreiter and Schmidhuber, 1997), use gating units (blue) to control information flow in and out of the internal state (gray), and angularization units (purple) to convert raw outputs into angles. Rightmost Inset: Angularization units select from a learned set of torsion angles (“alphabet”) a mixture of torsions, which are then averaged in a weighted manner to generate the final set of torsions. Mixing weights are determined by computational units.