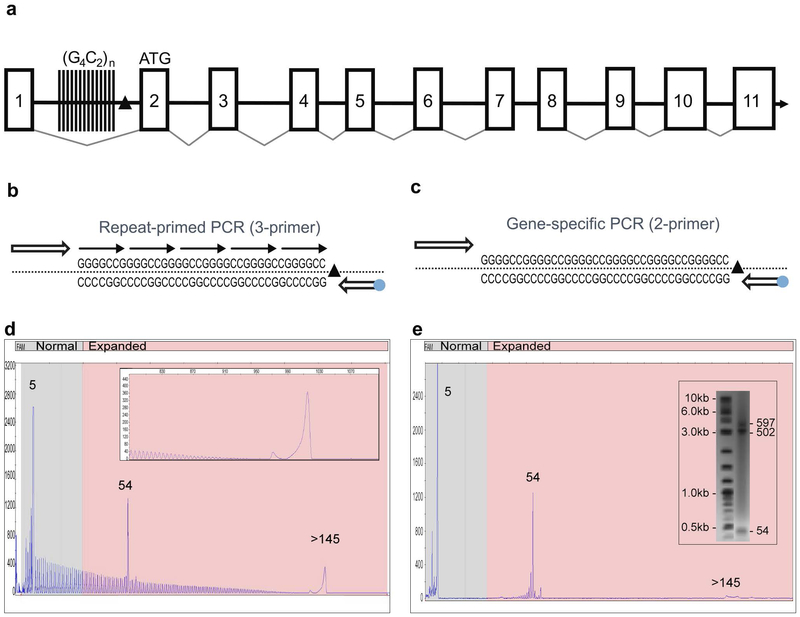
Figure 1: A novel PCR technology for the amplification of the hexanucleotide G4C2 repeat element in the C9orf72 gene.
A) Schematic representation of the C9orf72 gene structure showing the predicted 11 exons (boxes), the location of the intronic hexanucleotide repeat expansion (vertical lines) and the 3’SV region (black triangle). B) Schematics of the 3-primer GS/RP-PCR design (blue dot – FAM fluorophore) and C) 2-primer GS-PCR design. D) A representative 3-primer GS/RP-PCR/CE profile for an expanded sample (ND10206) that overlays GS peaks (a normal allele at 5 repeats, an expanded allele at 54 repeats) with a hyper-expanded RP-PCR profile and corresponding pile-up peak (inset, >145 repeats). E) Matching 2-primer GS-PCR products for Coriell sample ND10206 resolved on CE and AGE (inset) shows the two alleles (sized at 5 and 55 repeats). The hyper-expanded peak is resolved in the gel image as a 502 and 597 repeats.