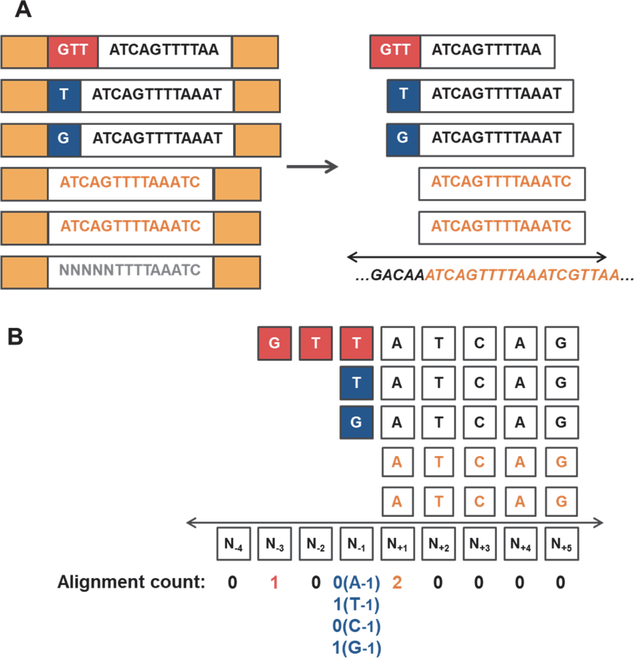
Figure 2. 5’-end sequence analysis pipeline to detect activities of Thg1/TLPs.
5’-addition activity could generate 5’-sequences that do not match genome-encoded nucleotides expected from incorporation by transcription. A) The pipeline developed here distinguishes between read sequences (shown in white boxes) that precisely match annotated mature RNA 5’-ends (shown in orange text) vs. read sequences that contain different nucleotides at the 5’-end, possibly added by Thg1/TLP enzymes (red/blue boxes). After removal of adapter sequences (solid yellow boxes), trimming, and quality filtering of reads (poor quality read indicated by gray text), 5’-end nucleotides that map upstream of the annotated mature RNA 5’-end are preserved by soft-clipping, which allows up to 10 nucleotides at the 5’-end that do not match the genomically-encoded nucleotide(s) at this location (genomic sequence is indicated under the double-arrow). B) Quantification of alignment count values for reads corresponding to a representative non-coding RNA. After reads are aligned, the number of reads with a 5’-end that corresponds to each of 10 nucleotides surrounding the annotated mature RNA 5’-end (N+1) are quantified. For position N−1, the reads were further analyzed according to the base identity (A, T, C, or G) at this location.