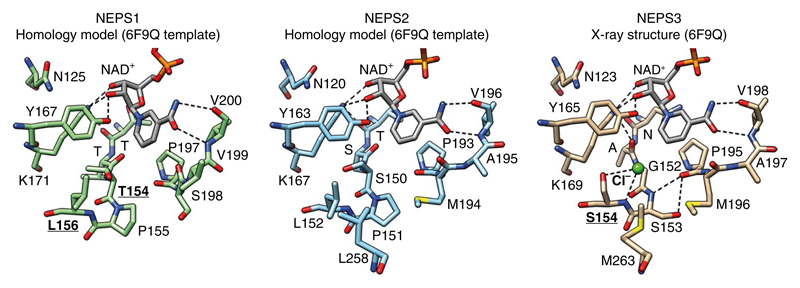
Fig. 5. Structure of NEPS enzymes.
X-ray crystal structure of NEPS3 (6F9Q) and homology model structures of NEPS1 and NEPS2. Active site NAD+ and residues are depicted as sticks. Dashed lines represent proposed hydrogen bonds. The NEPS3 active site lacks the characteristic Ser/Thr of the SDR catalytic tetrad (Gly152). It also features a chloride bound to Ser154 and hydrogen bonding between Ser153 and Pro190. Ser154 is replaced by leucine in NEPS1 (Leu156) and NEPS2 (Leu152). The role of residues that are boldface and underlined have been analyzed by mutation. See Supplementary Fig. 15 for further analysis of the NEPS3 crystal structure.