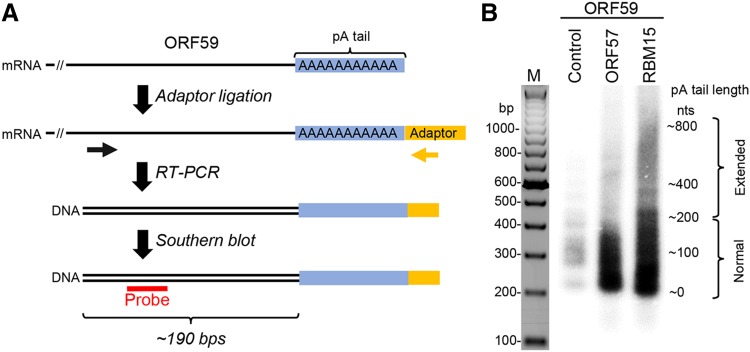
Fig. 6.
ORF57 prevents RBM15-mediated hyperpolyadenylation of KSHV ORF59 RNA. A A workflow of the strategy in determination of RNA poly (A) (pA) tail length (blue). First, a chimeric RNA–DNA adaptor (yellow, rUrUrUAACCGCGAATTCCAG/3AmM/-3′) was ligated to the mRNA 3′ end and followed by RT with an adaptor-specific antisene primer (5′-GACTAGCTGGAATTCGCGGTTAAA-3′). The cDNA was amplified by PCR using an ORF59-specific primer (black arrow, 5′-GGATCGTGGGAAGGTGCC-3′) in combination with an adaptor-specific primer (yellow arrow, 5′-GACTAGCTGGAATTCGCGGTTAAA-3′). The ORF59-specific primer is positioned approximately 190 bps upstream of the pA start (black double lines). The obtained PCR products (black double lines) are analyzed by Southern blot using an ORF59-specific probe (red, 5′-AATCAGGGGGTTAAATGTGGT-3′). B Length of the ORF59 RNA pA tail in HEK293T cells transfected with a vector expressing ORF59-FLAG fusion protein (pVM18) in the absence (control) or presence of ORF57 or RBM15 was determined by Southern blot as described in (A). Total RNA isolated from HEK293T cells was used in this study. The RT-PCR products of ORF59 RNA without a pA tail was about 190 bps in size and the varied sizes or smear signals of the ORF59 cDNA indicate heterogenic length of the pA tails.