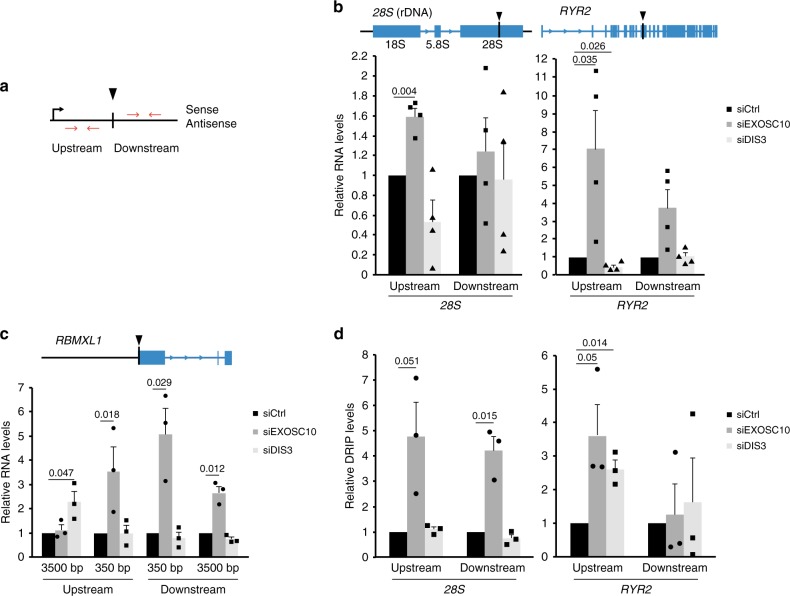
Fig. 4.
DilncRNA levels are increased upon depletion of EXOSC10. a Schematic drawing of a DSB showing the nomenclature used to describe the origin of dilncRNAs transcribed from DSB-flanking sequences. b SsRT-qPCR experiments were carried out to analyse the levels of upstream antisense and downstream sense (at 500 bp from the DSB) RNAs in the 28S and RYR2 genes in HeLa cells, as indicated, 20 h after transfection with the pOPRSVI/MCS-I-PpoI plasmid for I-PpoI expression. The graph shows the ratios cut/uncut for siEXOSC10 and siDIS3 samples, compared to the ratios observed in the control cells (siCtrl). RNA levels were normalized to ARPP. c DIvA cells were transfected with siCtrl, siEXOSC10, or siDIS3 and incubated for 4 h with 300 nM 4-OHT before RNA analysis. The graph shows relative RNA levels quantified by ssRT-qPCR as in b. d DRIP-qPCR was performed in HeLa cells 20 h after transfection with the pOPRSVI/MCS-I-PpoI plasmid. The graph shows the relative DRIP-qPCR levels in cells treated with either siEXOSC10 or siDIS3 compared to control cells (siCtrl). DRIP-qPCR levels were normalized to GADPH. Error bars show s.e.m. from four independent experiments in b and d, and three in c. Statistical testing was done using a one-sample Student’s t-test and significant p-values are shown in the figure. Source data for Fig. 2a–c are provided as a Source Data file